Abstract
The basic helix-loop-helix transcription factor Ascl1 plays a critical role in the intrinsic genetic program responsible for neuronal differentiation. Here, we describe a novel model system of P19 embryonic carcinoma cells with doxycycline-inducible expression of Ascl1. Microarray hybridization and real-time PCR showed that these cells demonstrated increased expression of many neuronal proteins in a time- and concentration-dependent manner. Interestingly, the gene encoding the cell cycle regulator Gadd45γ was increased earliest and to the greatest extent following Ascl1 induction. Here, we provide the first evidence identifying Gadd45γ as a direct transcriptional target of Ascl1. Transactivation and chromatin immunoprecipitation assays identified two E-box consensus sites within the Gadd45γ promoter necessary for Ascl1 regulation, and demonstrated that Ascl1 is bound to this region within the Gadd45γ promoter. Furthermore, we found that overexpression of Gadd45γ itself is sufficient to initiate some aspects of neuronal differentiation independent of Ascl1.
Keywords: Differentiation, Ascl1, Gadd45γ
Introduction
Transcription factors of the basic helix-loop-helix (bHLH) class play important roles in many aspects of neuronal development. The importance of bHLH genes for neurogenesis was first appreciated in Drosophila melanogaster, where it was shown that genes belonging to the achaete-scute complex are required for the development of some neurons in the peripheral and central nervous system (PNS and CNS; Romani et al., 1989; González et al., 1989). Genetic studies in Drosophila and Xenopus have also shown that bHLH proteins are both necessary and sufficient to commit ectodermal progenitors to a neuronal-specific fate, and that this activity involves the Notch signaling pathway (Turner and Weintraub, 1994; Artavanis-Tsakonas et al., 1999). The proneural function of bHLH genes appears to have been evolutionarily conserved: homologues of achaete-scute genes have been identified in a variety of vertebrate species, and these genes regulate the development of specific classes of neurons (Johnson et al., 1990; Guillemot et al., 1993). For example, mammalian achaete-scute homolog 1 (Ascl1) is expressed in subsets of proliferating precursor cells in the PNS and CNS of the mouse embryo, and knockout analysis has shown that Ascl1 is required for the development of autonomic neurons and olfactory receptor neurons (Guillemot et al., 1993). The neurogenic effects of bHLH proteins—such as Ascl1—make them useful in strategies to yield neuron-enriched grafts. Recently, transduction of Ascl1 into donor neuronal progenitor cells before transplantation dramatically enhanced neuronal yield and donor cell survival, both in vitro and in vivo (Yi et al., 2008).
The function of the vertebrate CNS is dependent on the generation of neuronal progenitor cells at the proper developmental time, making the balance between proliferation and cell cycle withdrawal fundamental to the formation of the mature vertebrate CNS. Proneural bHLH proteins promote cell cycle arrest, presumably through activation of cyclin-dependent kinase inhibitors (Farah et al., 2000). Despite the importance of neurogenic bHLH families in neuronal development, primary target genes and transcriptional programs directly regulated by neurogenic bHLH proteins have yet to be systematically defined.
P19 cells are pluripotent embryonic carcinoma (EC) cells that differentiate into cell types of all three germ layers (McBurney et al., 1982), and are a commonly used model to study neuronal differentiation in vitro. Treatment of P19 cells with retinoic acid followed by aggregation results in neuronal and glial differentiation (Bain et al., 1996). Many bHLH genes are induced in this method of differentiation, including Ascl1, and its pattern of expression closely matches those observed in vivo (Johnson et al., 1992). More recently, transient transfection of neural bHLH proteins such as Ascl1 was shown to be sufficient to convert P19 cells into a relatively homogeneous population of electrophysiologically differentiated neurons (Farah et al., 2000). These findings suggest that undifferentiated P19 cells express the genes necessary to support the initiation of neuronal differentiation in response to neurogenic bHLH transcription factors. One limitation to the current studies of Ascl1-induced neuronal differentiation is their reliance on transient transfection, which results in difficulty controlling Ascl1 expression temporally or quantitatively. Furthermore, the levels of transfected DNA are heterogeneous at a cellular level. To circumvent these problems, we developed an inducible P19 cell line in which the expression of the Ascl1 gene was under the control of the tetracycline transcriptional repressor (Gossen and Bujard, 1992).
In our studies, we used microarray hybridization analysis combined with tetracycline-regulated Ascl1-expressing cell lines to delineate the transcriptional consequences of Ascl1 induction. We showed that doxycycline induction of Ascl1 in P19 cells caused expression of neuronal marker proteins, including cytoskeletal and synaptic proteins, in a time- and dose-dependent manner and generated neurons that were polarized and electrically excitable. Microarray analysis of genes induced over the time course of differentiation showed changes in several genes not previously characterized as Ascl1 responsive in P19 cells. One highly induced gene, growth-arrest and DNA-damage inducible protein 45 gamma (Gadd45γ), was of particular interest because of its role in cell cycle regulation (Smith et al., 1994; Wang et al., 1999; Zhan et al., 1999; Yang et al., 2000). Using reporter constructs of the human Gadd45γ gene that contained four evolutionarily conserved E-box consensus sites adjacent to the Gadd45γ promoter, we showed transactivation of Gadd45γ with Ascl1 in P19 cells. Additionally, chromatin immunoprecipitation (ChIP) assays showed that Ascl1 associates with the Gadd45γ promoter in living P19 cells, supporting our data that Gadd45γ is a direct transcriptional target of Ascl1. Finally, using a Gadd45γ-inducible P19 cell line, we found that overexpression of Gadd45γ recapitulated a subset of Ascl1-mediated gene regulatory events.
Experimental Methods
Materials
The following primary antibodies were used in the experiments: TetR, Tau (Chemicon), GAPDH, Map2 (Cell Signaling Technology), Ascl1 (BD Pharmingen), TuJ1 (Covance), Gap43 (Sigma-Aldrich), Isl1 (DSHB University of Iowa), Synaptophysin (Syp; BD Biosciences), and Gadd45γ (Sigma-Aldrich). Secondary horseradish peroxidase-conjugated antibodies were obtained from Cell Signaling Technology. Alexa Fluor conjugated antibodies (goat anti-mouse Alexa Fluor 488, goat anti-mouse Alexa Fluor 546, and goat anti-rabbit Alexa Fluor 488) were all purchased from Invitrogen. Five different shRNAs were assayed for efficacy of knocking down the mouse Gadd45γ gene and were obtained from Open Biosystems.
Cell culture, transfection, and treatment
P19 EC cells were cultured in Minimal Essential Medium Alpha (MEMα; Gibco) supplemented with 7.5% calf serum (CS; HyClone), 2.5% fetal bovine serum (FBS; HyClone), and 1% penicillin/streptomycin (Gibco). The Ascl1-inducible P19 cell line (P19T1A2) was maintained in the same media as P19 EC cells, with the addition of G418 (200 μg/ml, HyClone) and hygromycin (100 μg/ml, Invitrogen). HEK-293T cells were grown in Dulbecco's Modified Eagle Medium (Gibco) supplemented with 10% FBS. Cells were kept at a temperature of 37°C, a minimum relative humidity of 95%, and an atmosphere of 5% CO2 in air. Cells were maintained below 80% confluence and passaged by dissociating them into single cells using TrypLE Express (Gibco). Cells were transfected using the TransIT-LT1 transfection reagent (Mirus) following the manufacturer's instructions. When necessary, the appropriate parental expression plasmid DNA was added to maintain a constant total amount of DNA.
Construction of the Dox-controlled Ascl1 expression system
Initially, the pCMV-TetOnAdv or pUS2-TetOn plasmids were used to generate Doxycycline (Dox, a tetracycline derivative) inducible P19 cells. However, these attempts were largely unsuccessful. Therefore, we constructed the pUS2-TetOnAdv plasmid by subcloning the 1325 bp SalI/EcoRI fragment of pUS2 into XhoI/EcoRI digested pTetOnAdv (Clontech). Using the TransIT-LT1 reagent, P19 cells were transfected with the pUS2-TetOnAdv vector together with a 10-fold lower amount of the 2.2 kb BamHI fragment of pCMV-Neo containing the neomycin phosphotransferase gene under the control of the SV40 promoter. Stable clones were selected with 200 μg/ml of G418. G418-resistant colonies were screened by transient co-transfection with the pTRE-Luciferase vector (pTRE-Luc, Clontech), which encodes for the firefly luciferase protein under control of the TRE promoter, and RL-SV40 (Promega), which encodes for renilla luciferase protein and served as a control for transfection efficiency. A Dual Luciferase Reporter Assay (Promega) was used to identify clones exhibiting low background and high luciferase activity upon addition of Dox. The cell line used in further experiments was designated P19T1.
The pTRE-Ascl1 expression vector was constructed by subcloning the 700 bp EcoRI/XbaI fragment encoding Ascl1 from pCS2-Ascl1, and ligating this fragment with EcoRI/XbaI digested pTRE-tight (Clontech). P19T1 cells were co-transfected with pTRE-Ascl1 together with pTK-Hyg (Clontech), and cultured in the presence of 100 μg/ml hygromycin for selection. Hygromycin-resistant colonies were screened for the expression of neuron-specific class III β-tubulin (TuJ1) and Ascl1 upon addition of Dox by immunocytochemistry and western blot analysis.
Construction of the Dox-controlled Gadd45γ expression system
The pTRE-Gadd45γ-IRES2-EGFP vector was constructed by first inserting the 1.4 kb NheI/NotI fragment of pIRES2-EGFP (Clontech) into NheI/NotI digested pTRE-tight (Clontech) to generate the plasmid pTRE-tight-IRES2-EGFP. pUS2-Ascl1 was then digested with EcoRI and SnaBI. The resulting 760 bp fragment was ligated into EcoRI/SmaI digested pTRE-tight-IRES2-EGFP to generate pTRE-Ascl1-IRES2-EGFP. Finally, the human Gadd45γ coding region was PCR amplified from pCMV6-XL5-hGadd45γ (Origene) using the primer pair shown in Supplementary Table 1. The resulting 500 bp amplification product was purified, digested with EcoRI/XbaI, and ligated to EcoRI/XbaI digested pTRE-Ascl1-IRES2-EGFP, thereby replacing the Ascl1 coding sequence with the Gadd45γ coding sequence. P19T3 cells were transfected with 26 μg of pTRE-Gadd45γ-IRES2-EGFP and 1 μg of pUS2-puro. Puromycin resistant clones were identified by enhanced green fluorescent protein (EGFP) fluorescence after 24 h of doxycycline treatment and expanded. The clone P19T3GIE2 was chosen for detailed characterization based on the high induction of EGFP fluorescence.
Differentiation of P19T1A2 cells
For differentiation of P19T1A2 cells, tissue culture plates were laminin coated using a procedure adapted from Ray et al. (1995). Briefly, plates were coated in a solution of 5 μg/ml laminin (Invitrogen) diluted in phosphate buffered saline (PBS, HyClone). The plates were sealed in plastic bags and kept in an incubator overnight (37°C, 5% CO2). After aspirating off the laminin solution, the plates were washed twice with PBS before seeding the P19T1A2 cells at a density of 5.0 ×105 cells/ml. For the first four days of differentiation, cells were maintained in MEMα supplemented with 7.5% CS, 2.5% FBS, 1% penicillin/streptomycin, 200 μg/ml G418, 100 μg/ml hygromycin, and 0.5 μg/ml doxycycline. On day four, the media was changed to Neurobasal media (Gibco) supplemented with 1% penicillin/streptomycin, B27 (Gibco), GlutaMAX (Invitrogen), G418 (200 μg/ml), hygromycin (100 μg/ml), and doxycycline (0.5 μg/ml).
Construction of reporter plasmids
The pEL2 reporter vector containing the EGFP coding region fused to the firefly luciferase coding region was constructed to monitor reporter expression in living cells (EGFP) as well as to quantitate reporter expression by enzymatic assay (firefly luciferase). pEL2 was constructed by ligating the PCR-amplified coding region of firefly luciferase generated using pGL3basic (Promega) as template and the primer pairs shown in Supplementary Table 1. The resulting PCR fragment was Acc65I/NotI digested prior to subcloning into BsrBI/NotI digested pEGFP-1 (Clontech).
Oligonucleotides used in generating the following reporter plasmids can be found in Supplementary Table 1. The 1222 bp promoter sequence for the human Gadd45γ gene was PCR amplified from human genomic DNA (Clontech), and subcloned into pEL2 (1222-EL2). The AVID alignment program implemented in VISTA was used to compare conserved regions between human and mouse, and four E-box (CANNTG) sequences were identified clustered in a highly conserved region of the promoter of the human Gadd45γ gene. PCR procedures were used to generate five stepwise deletion constructs (938-, 665-, 281-, 194-, and 188-EL2) of the full length Gadd45γ promoter (1222-EL2). The amplified PCR fragments were subcloned into the pGEM-T-Easy vector system (Promega). The DNA was HindIII/BamHI digested and then subcloned into HindIII/BamHI digested pEL2. A 281-EL2 construct harboring substitutions to the two proximal E-box consensus sites was constructed by oligo-directed mutagenesis and PCR. The E-box consensus at -281/-275 was mutated from CACGTG to GAATTC, and the consensus at -194/-188 was mutated from CAGCTG to ACGCGT. Constructs containing the two mutated sites were generated in separate rounds of PCR. The PCR amplicon containing the distal E-box mutation was EcoRI/MluI digested. The amplicon containing the proximal E-box mutation was MluI/BamHI digested. These fragments were then ligated into EcoRI/BamHI digested pEL2. All of the Gadd45γ deletion subclones were sequenced to ensure that only the intended deletions were introduced. All oligonucleotides were synthesized by Invitrogen.
Electrophysiological recordings
All recordings were carried out at room temperature using an external solution that contained (in mM) 132 NaCl, 5.3 KCl, 1.3 NaH2PO4, 1.7 MgSO4, 5.4 CaCl2, 12 Hepes, 6.3 glucose, pH, 7.4. Whole-cell recordings on P19T1A2 cells treated with Dox for six days were made using a Dagan 3900A amplifier in bridge mode. Neurons were visualized with an Olympus BX51WI upright microscope equipped with differential interference contrast optics. Patch-pipettes made from Clark Borosilicate Standard Wall glass (Warner Instruments) and pulled using a P-97 Flaming-Brown pipette puller (Sutter Instruments) with resistances of 9-11 MΩ were used and filled with the following internal solution (in mM): KCl 140, NaCl 5, MgCl2 1, Na2EGTA 10, Hepes 10, pH 7.4. Seal resistances of >2 GΩ were achieved prior to rupturing into whole-cell mode. Action potentials were elicited by delivering a 5 ms current step of increasing amplitude (0.01 nA steps).
Microarray analysis
Total RNA (200 ng) was amplified and labeled using the Illumina Total Prep RNA Amplification Kit (Ambion). Labeled cRNA (1.5 μg) was hybridized at 55°C for 22 h to Sentrix-6 Mouse V1.0 BeadChip microarrays (Illumina). Microarrays were washed and scanned for data collection as directed by the manufacturer. Microarray data were analyzed using BeadStudio software (Illumina). Differential gene expression was determined using quantile normalization and the Illumina Custom error model. mRNAs for analysis were selected based on mRNAs detected in at least one condition with p < 0.01. For differential expression analyses, a cutoff of p < 0.01 was used. All analyses used a subset of Illumina probes that matched sequences in the Refseq database and mapped to the mouse genome at a single location (Pinglang Wang and Fan Meng, University of Michigan, personal communication).
RNA isolation and RT-PCR analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) in accordance with the manufacturer's instructions. Single stranded cDNA was synthesized from 2 μg of total RNA using SuperScript II Reverse Transcriptase and random hexamers (Invitrogen). Gene expression was evaluated by real-time quantitative PCR (RT-PCR) using the SYBR Green PCR Master Mix (Applied Biosystems), and the MyiQ single-color real-time PCR detection system (Bio-Rad) according to manufacturer's instructions. The specificity of the PCR amplification procedures was checked with a heat dissociation protocol (from 72°C to 98°C) after the final cycle of the PCR. Each reaction was done in triplicate. Expression levels were calculated using the delta-delta CT method, with GAPDH as the normalization control. The primer pairs used to amplify target genes are shown in Supplementary Table 1.
Dual luciferase reporter assay
Dual luciferase assays were performed using the Dual Luciferase Reporter Assay (Promega) following recommended protocols. Samples were read on a FLUOstar OPTIMA microplate reader (BMG Labtech). To correct for differences in transfection efficiencies, firefly luciferase activity (pEL2) was normalized to that of renilla luciferase (pUS2-RL). The pUS2-RL plasmid was constructed by subcloning the 1.3 kb BglII/XbaI fragment of pUS2 containing the Ubc promoter into BglII/NheI digested pRL-SV40. Experiments were repeated a minimum of three times and results were expressed as mean ± standard deviation. The statistical significance of Ascl1 transactivation data was determined by employing a student's paired t-test (p < 0.01).
Chromatin immunoprecipitation assay
P19T1A2 cells were treated with 5 μg/ml Dox for 24 h as described above. An antibody against Ascl1 was used for immunoprecipitation (BD Pharmingen) and the chromatin immunoprecipitation (ChIP) assay was performed as described by the manufacturer (Cell Signaling). The immunoprecipitates were subjected to RT-PCR using primers specific to the Gadd45γ promoter. The resulting amplified fragment contained both the E3 and E4 E-boxes of the Gadd45γ promoter. The ChIP amplifications were performed using SYBR Green PCR Master Mix (Applied Biosystems) in quadruplicate. The reaction conditions were as follows: 95°C for 7 min, followed by 60 cycles of 95°C for 15 sec, 55°C for 15 sec and 72°C for 20 sec. Threshold cycle numbers (CT) were determined with the MyiQ single-color real-time PCR detection system (Bio-Rad). The DNA levels from the ChIP RT-PCR assay were calculated using the delta-delta CT method, with primers for the ribosomal protein L30 (RPL30) as the normalization control. PCR primer sets for the ChIP assays are shown in Supplementary Table 1.
SDS-PAGE and western blot analysis
Cells were washed twice with Dulbecco's PBS (DPBS; Hyclone) and lysed in buffer containing 10 mM NaH2PO4•H2O, 1 mM EDTA, 1 mM DTT, 250 mM sucrose, 10 mM sodium fluoride, complete EDTA-free protease inhibitors (Roche), and 1 mM PMSF. Lysates were sonicated, and protein concentrations were determined by the bicinchonic acid protein assay (Bio-Rad). Equal amounts of total protein were denatured at 95°C in the presence of SDS and β-mercaptoethanol. Samples were resolved on linear gradient Tris-HCl gels (Bio-Rad) and transferred onto 0.2 μm polyvinylidene difluoride membranes. Detection was carried out using Lumi-Light Western Blotting Substrate (Roche) according to the manufacturer's instructions.
For resolving Gadd45γ protein in P19T1A2 cells, samples were transferred to 0.2-μm nitrocellulose membranes (BA-83, Whatman). Membranes were blocked for 4 h in PBS supplemented with 5% non-fat dried milk, 2% polyvinylpyrrolidone (PVP-40), and 0.1% Triton X-100 and subsequently incubated with a 1:200 dilution of anti-Gadd45γ in PBS supplemented with 0.5% bovine serum albumin and 0.1% Triton X-100 overnight at 4°C. Membranes were washed three times for 10 min with TBST (50 mm Tris, pH 7.5, 150 mm NaCl, and 0.05% Tween 20), and then incubated with a 1:2,000 dilution of goat anti-mouse-HRP (Cell Signaling Technology) in TBST supplemented with 5% non-fat dried milk as the secondary antibody for 2 h. Following the final set of three 10 min washes with TBST, the blots were developed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) according to the manufacturer's instructions.
Recombinant His-tagged Gadd45γ protein was purified from E. coli essentially as described (Collins and Uhler, 1999), and the purified Gadd45γ protein had an apparent molecular weight of 17 kDa on SDS-PAGE. Quantitative assay of antigen expression was based on density measurements of protein bands using ImageJ software (http://rsb.info.nih.gov/ij).
Immunocytochemistry
Cells were washed twice with DPBS, and then fixed in 4% formaldehyde solution for 10 minutes. Cells were washed twice in PBS, and then blocked for one hour in PBS supplemented with 2% goat serum and 0.1% Triton X-100. Cells were probed with primary antibodies diluted in blocking solution for two hours at 23°C. After washing in PBS, cells were incubated with AlexaFluor conjugated secondary antibodies for one hour at 23°C, followed by three PBS washes. For nuclear counterstaining, the cells were incubated in 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen) for 10 minutes before being washed twice in PBS and visualized. To collect still images, we used an inverted Olympus IX70 fluorescence microscope using an Illix CCD imaging system and Micro Computer Image Device software (Imaging Research Inc.). Confocal images were obtained using an inverted Olympus FV1000 laser scanning confocal microscope. Prior to image collection, the acquisition parameters for each channel were optimized to ensure a dynamic signal range and to ensure no signal bleed through between detection channels.
Results
Generation of rtTA-expressing clones derived from P19 cells
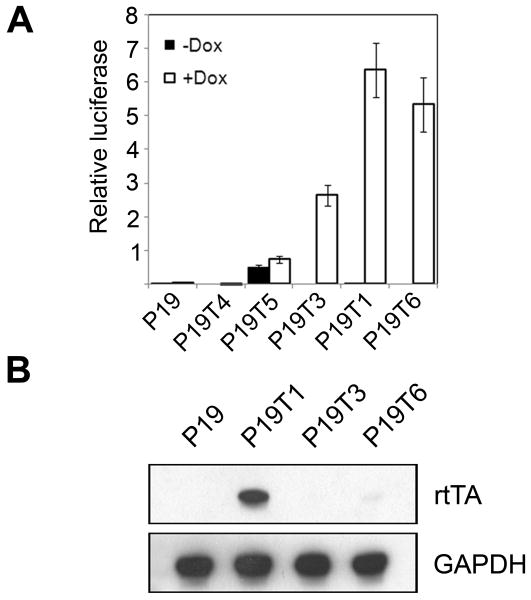
A total of 156 putative rtTA-stable clones were screened by transfection with the reporter plasmid pTRE-Luc and grown with or without doxycycline (Dox) in the medium. The majority of the clones showed no regulation of firefly luciferase activity (e.g. P19T4; Figure 1A). Six clones showed high constitutive firefly luciferase activity in the absence of Dox (e.g. P19T5), and six clones showed high firefly luciferase activity only in the presence of Dox (e.g. P19T3, P19T1, and P19T6). Clones P19T1, P19T3, and P19T6 showed the highest induction of luciferase activity, exhibiting 212-, 263-, and 532-fold increases in the presence of Dox, respectively. Upon passage, the induction by Dox in the P19T3 and P19T6 cells progressively diminished. However, the P19T1 cells showed consistent induction over 20-30 passages. After extended passages, induction of pTRE-Luc activity was roughly correlated with the amount of rtTA protein expressed in the cells. Figure 1B shows that clone P19T1, which had the highest sustained induction of luciferase activity in the presence of Dox, also expressed the most rtTA protein. We selected the P19T1 clone for the generation of secondary transfectants in subsequent experiments.
Figure 1. Generation of rtTA-stable P19 cells.
A. Relative luciferase activity in five putative P19 clones stably transfected with the rtTA expression plasmid pUS2-TetOnAdv. Compared to the negative control of wild-type P19 cells, clones P19T3, P19T1, and P19T6 showed significant induction of luciferase activity, but only P19T1 showed sustained induction after multiple passages. Results are shown as mean firefly luciferase expression levels relative to renilla luciferase controls ± s.d. B. Expression of the rtTA protein in the three P19 clones that showed strong Dox regulation of luciferase activity in (A). Western blot analysis (50 μg of protein/lane) using an antibody against rtTA showed that clone P19T1, which had the highest sustained induction of luciferase activity in the presence of Dox, also expressed the most protein.
Generation of a stably transfected cell line showing Dox-responsive Ascl1 expression
P19T1 clones stably co-transfected with plasmids pTRE-Ascl1 and pTK-hygro were generated as described in Materials and Methods. Of the 206 hygromycin-resistant clones isolated, six showed a significant reduction of growth in the presence of Dox. Microtubule-associated protein 2 (Map2) and neuron-specific class III β-tubulin (TuJ1) are widely accepted as neuronal marker proteins, and are induced in P19 cells transiently transfected with Ascl1 (Farah et al., 2000). Immunocytochemistry was performed with Map2 and TuJ1 antibodies. Three pTRE-Ascl1 transfected clones—P19T1A2, P19T1A3, and P19T1A12—produced a high percentage of cells (>30%) that were Map2- and TuJ1-immunoreactive in the presence of Dox, although their levels of Ascl1 expression varied (data not shown). This induction of Map2 and TuJ1 immunoreactivity was never observed following treatment of parental P19 or P19T1 cells with Dox (data not shown). We chose the P19T1A2 clone for subsequent experiments because of the low level of spontaneous differentiation and the high level of differentiation in the presence of Dox.
Optimizing growth conditions for Ascl1-induced neuronal differentiation in P19 cells
Neuronal differentiation and survival in vivo and in vitro depends on a variety of factors. Pure neuronal cultures require specific growth factors for optimal survival and neurite production. In addition to these soluble factors, the culture substrate is essential for neuronal adhesion and influences the number, shape, and growth rate of neurites (Rogers et al., 1983). Experiments were performed to define the relative importance of these various influences and optimize growth conditions for our model of neuronal differentiation.
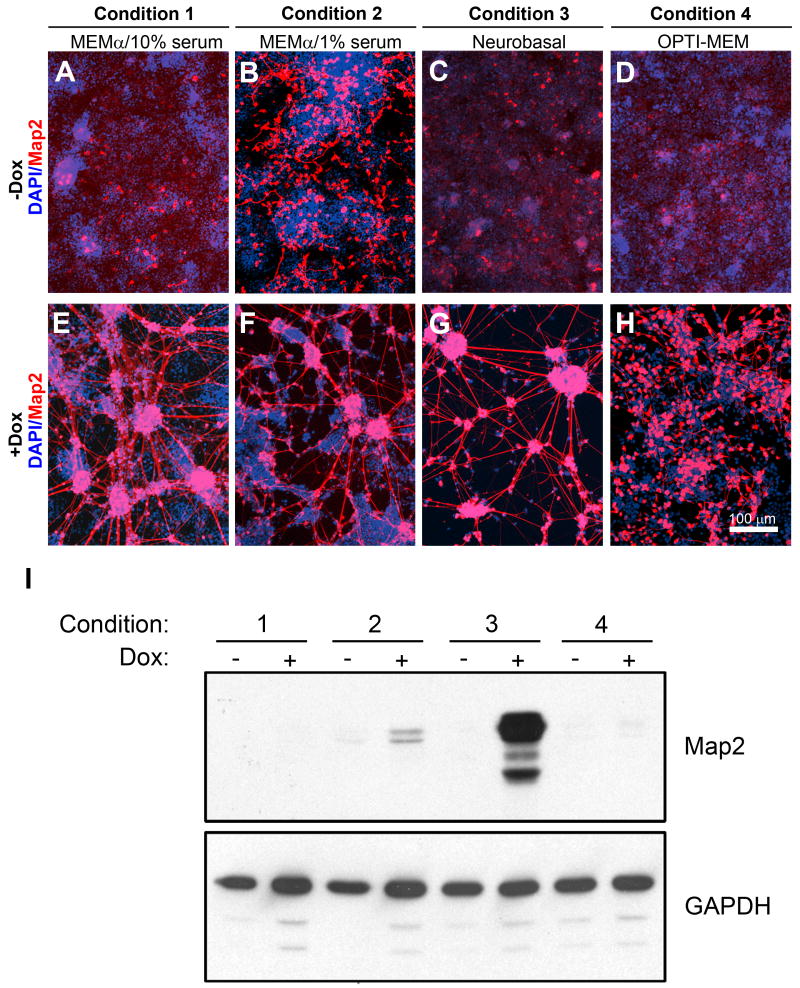
Substrata commonly used for neuronal cell culture include polymers of basic amino acids such as poly-D-lysine and polyornithine, and extracellular matrix constituents such as collagen, fibronectin, and laminin (Carbonetto et al., 1983; Lochter and Schachner, 1993). Three substrata were evaluated: poly-D-lysine, poly-ornithine, and laminin. Neurite outgrowth was consistently enhanced in the presence of laminin (data not shown). P19T1A2 cells were seeded onto laminin-coated tissue culture plates and treated with or without Dox using four different growth conditions: 1. MEMα (7.5% CS, 2.5% FBS; Figure 2A, E), 2. MEMα (1% FBS; Figure 2B, F), 3. MEMα (7.5% CS, 2.5% FBS) for the first three days of differentiation, followed by a change to Neurobasal media (B27, GlutaMAX) for the remaining duration of the differentiation protocol (Figure 2C, G), and 4. OPTI-MEM (1% FBS; Figure 2D, H).
Figure 2. Optimization of growth conditions for differentiation.
A-H. Map2 (red) staining of P19T1A2 cells treated ± Dox at 0.5 μg/ml for 10 days under the following growth conditions: (1) MEMα (7.5% CS, 2.5% FBS), (2) MEMα (1% FBS), (3) MEMα (7.5% CS, 2.5% FBS) for the first three days of differentiation, followed by a switch to Neurobasal media (B27, GlutaMAX), and (4) OPTI-MEM (1% FBS). Nuclei were visualized with DAPI and appear blue. Under the first three growth conditions, cells differentiated into neurons that were immunoreactive to Map2 in the presence of Dox (E-G), although cells grown in reduced serum were immunoreactive to Map2 protein in the absence of Dox (B). Cells grown in OPTI-MEM resulted in Map2-positive cells that were morphologically less differentiated than the other conditions (H). Scale bar = 100 μm. I. Western blotting for expression of Map2 protein under the growth conditions shown in (A-H). The highest amount of Map2 protein was observed in cells cultivated in Neurobasal media (Condition 3).
At ten days and for all media conditions, more P19 cells adopted a neuronal morphology and expressed the appropriate neuronal-specific markers—such as Map2—in the presence of Dox than in the absence of Dox (Figure 2E-H). However, the Map2-positive cells cultured in OPTI-MEM (Condition 4) appeared less differentiated than in other conditions as evidenced by a lack of neurites (Figure 2H). Cells grown under reduced (1% FBS) serum conditions also expressed significant Map2 in the absence of Dox, although the expression was not found in long neurites (Figure 2B). This observation is consistent with previous reports suggest that cultivating EC cells in a low serum environment can cause them to spontaneously differentiate into neurons (Pachernik et al., 2005).
Changing culture media from MEMα to Neurobasal media on the fourth day following Dox treatment (Condition 3) was chosen as the optimal growing condition because it resulted in the highest percentage of Map2-positive cells in the presence of Dox (Figure 2G), while few cells (< 0.1%) expressed Map2 in the absence of Dox (Figure 2C). Compared to other commonly used media (e.g. MEMα and DMEM), Neurobasal media has been shown to select against the proliferation of glia and increase neuronal viability (Brewer et al., 1993). Supporting these data, cells differentiated using Neurobasal media (Condition 3) also expressed the highest levels of Map2 protein when quantitated via western blot analysis (Figure 2I). Because equal amounts of protein are loaded on the western blot, contributions of undifferentiated cells (seen as blue nuclei in Figures 2E, 2F and 2H) to the total protein significantly dilute the Map2 signal seen on the western blot. Similar immunocytochemistry and western blot experiments with two additional P19 clones showing dox-inducible expression of Ascl1 (designated P19T1A3 and P19T1A12) also demonstrated that Condition 3 was optimal for neuronal differentiation (data not shown).
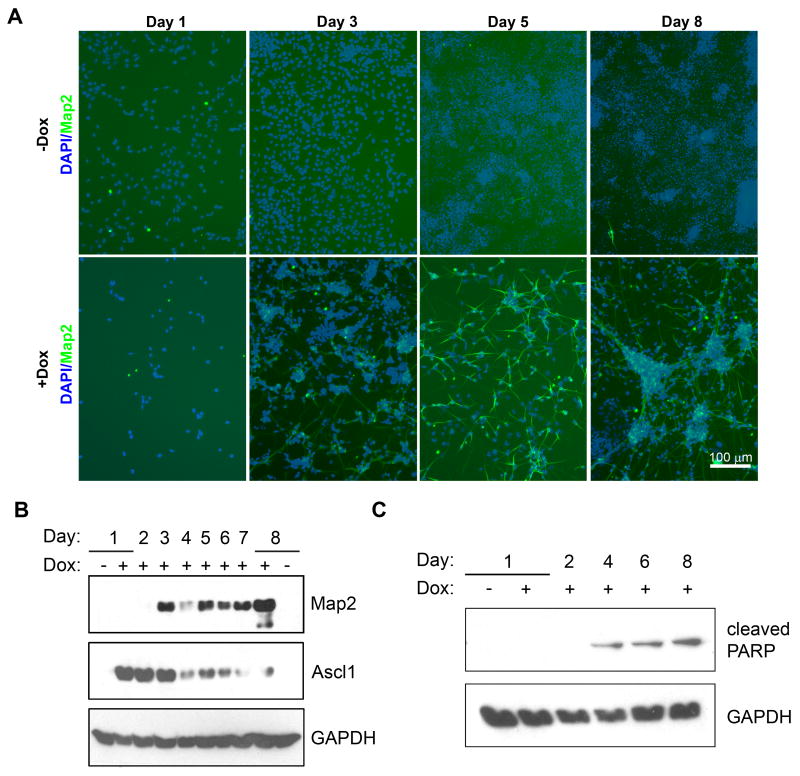
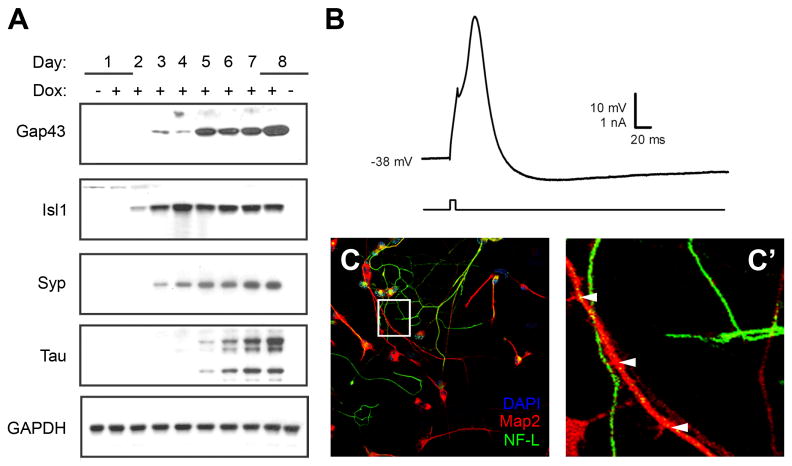
Under the growth conditions delineated above, a time course of differentiation with the P19T1A2 cells was carried out. Map2-positive cells first appeared three days after induction of Ascl1. Their total number increased progressively during the time course of differentiation, and the cells adopted a neuronal morphology (Figure 3A). Ascl1 protein was detected as early as one day after Dox treatment, remained elevated until three days, and then declined over the remaining four days of differentiation (Figure 3B). This transient induction of TRE-driven expression vectors has been observed with other P19T1 derived cells and it is possible that the transactivation by the TetOn-VP16 fusion protein is down regulated during differentiation of the P19 cells. We attribute at least a portion of the transient down-regulation of Map2 expression at day four to neuronal atrophy prior to the Neurobasal media change that offers the best trophic support (see Figure 2C). Neuronal atrophy was further confirmed by our examination of a second major neuronal marker, β-III-tubulin, whose expression was also transiently reduced prior to the switch to Neurobasal media (data not shown). Supporting these results, we examined the occurrence of apoptosis in P19T1A2 cells and observed expression of cleaved PARP—an apoptotic marker—beginning at four days of differentiation (Figure 3C).
Figure 3. Time course of differentiation with P19T1A2 cells.
A. Map2 (green) staining of P19T1A2 cells treated ± Dox at 0.5 μg/ml for the indicated days. Nuclei were visualized with DAPI and appear blue. In the absence of Dox, no Map2-positive cells were observed. In the presence of Dox, Map2-positive cells became evident after three days of treatment with Dox. Cells expressing Map2 underwent neuritogenesis by day five, and by day eight, the somas of Map2-positive cells began to cluster together, while the neurites became elongated and better defined. Scale bar = 100 μm. B. Western blot for expression of Map2 and Ascl1 protein during the time course of differentiation of P19T1A2 cells. C. Western blot for expression of cleaved PARP protein in P19T1A2 cells.
P19T1A2 cells respond to Dox in a dose-dependent manner
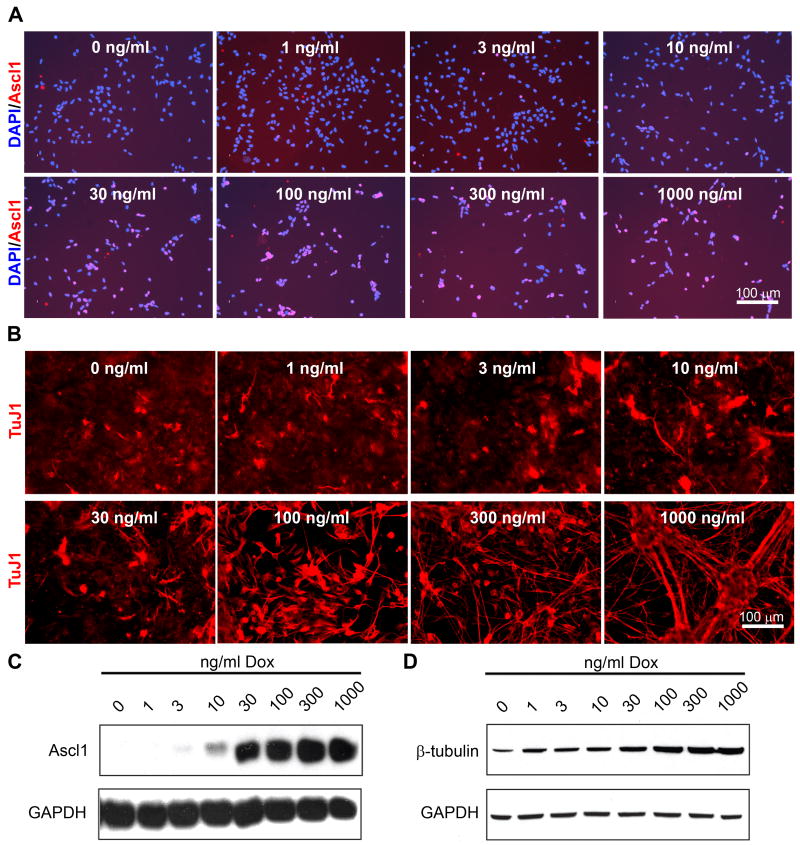
P19T1A2 cells were exposed to increasing concentrations of Dox (0, 1, 3, 10, 30, 100, 300, and 1000 ng/ml) in culture medium. Immunocytochemistry of P19T1A2 cells treated with varying concentrations of Dox for 24 h showed that individual Ascl1-positive cells became evident at a minimal concentration of 30 ng/ml of Dox, and that the percentage of Ascl1-positive cells increased with higher concentrations of Dox (Figure 4A). Ascl1 protein expression by western blot analysis was detectable in cells treated with as low as 3 ng/ml Dox, and was fully induced at 100 ng/ml Dox (Figure 4C). When P19T1A2 cells were treated with these same Dox concentrations for eight days, TuJ1 immunoreactivity also increased in a dose-dependent manner (Figure 4B). The shortest time to result in maximal TuJ1 staining was determined to be eight days (data not shown). Additionally, cells exposed to higher concentrations of Dox adopted a neuronal morphology and had an overall reduction in the density of the cells. The most striking effect was seen at Dox concentrations of 100 ng/ml or more where clustering of cell bodies and fasciculation of the neurite-like processes was observed. Western blot analysis verified the dose-dependent increase in β-III-tubulin protein expression (Figure 4D).
Figure 4. P19T1A2 cells express Ascl1 and differentiate in response to Dox in a dose-dependent manner.
A. Ascl1 (red) staining of P19T1A2 cells treated with the indicated concentrations of Dox for 24 h. Treatment of P19T1A2 cells with as low as 3 ng/ml of Dox resulted in sporadic Ascl1-positive cells, and the number and intensity of Ascl1-positive cells increased with greater concentrations of Dox. B. TuJ1 (red) images P19T1A2 cells treated with the indicated concentrations of Dox. P19T1A2 cells expressed TuJ1 in a dose-dependent manner. Furthermore, in the highest concentrations of Dox, the total number of cells was reduced due to the inhibition of proliferation resulting from neuronal differentiation. Scale bars = 100 μm. C, D. Western blot analysis for expression of Ascl1 (C) and β-III-tubulin (D) protein showed that protein expression correlates with the results shown in (A and B).
Identification of gene expression changes during Ascl1-induced neuronal differentiation
In order to determine the transcriptional profiles of P19T1A2 cells undergoing neuronal differentiation in vitro, we utilized microarray hybridization to characterize genes that were differentially expressed in P19T1A2 cells following Ascl1 induction. The abundance of mRNAs for over 270 known genes was induced four-fold or greater after eight days of differentiation, and the abundance of mRNA for over 80 genes was reduced by four-fold or greater (data not shown). The identities, associated functions, and mRNA fold changes of some key genes are provided in Table 1. Multiple values for fold changes represent data generated from distinct probes within the microarray. Many embryonic stem cell markers such as Pou5F1 (also known as Oct 3/4) were observed to decrease in the microarray hybridization. While several of the genes have previously been shown to be Ascl1 regulated, many have not previously been reported to be Ascl1 responsive (e.g. Npy, Fgf5, and Igf2).
Table 1. Selected gene expression changes by microarray analysis at eight days following Ascl1 induction.
| Gene Symbol | Gene Name(s) | Function/Process | Fold Change(s) |
|---|---|---|---|
| Igfbp5 | Insulin-like growth factor binding protein 5 | Growth factor binding | (75, 59, 59) |
| Dner | Delta/notch-like EGF-related receptor | Notch binding | (65) |
| Mapt | Microtubule-associated protein tau | Negative regulation of microtubule depolymerization | (35) |
| Isl1 | ISL1 transcription factor | DNA binding; neuron differentiation | (31) |
| Chgb | Chromogranin B | Protein binding | (27) |
| Map2 | Microtubule-associated protein 2 | Cytoskeletal regulatory protein binding | (24) |
| Igf2 | Insulin-like growth factor 2 | Growth factor activity | (23) |
| Syp | Synaptophysin | Syntaxin-1 binding; synaptic transmission | (22) |
| Slc17A6 | Solute carrier family 17; Vglut2 | L-glutamate transmembrane transporter activity | (19, 18) |
| Svop | SV2 related protein | Ion transmembrane transporter activity | (19) |
| Syt4 | Synaptotagmin IV | Calcium ion binding; neurotransmitter secretion | (17) |
| Gap43 | Growth associated protein 43 | Calmodulin binding; axon guidance | (7.5) |
| Dnmt3b | DNA methyltransferase 3B | DNA (cytosine-5-)-methyltransferase activity | (0.29, 0.15) |
| Folr1 | Folate receptor 1 | Folic acid transporter activity | (0.19, 0.11) |
| Pou5f1 | POU domain, class 5, transcription factor 1 | DNA binding; cell fate commitment | (0.13, 0.10) |
| FGF5 | Fibroblast growth factor 5 | Growth factor activity; cell proliferation | (0.11) |
| Lefty | Left right determination factor 1 | Growth factor activity; anterior/posterior axis specification | (0.11) |
| Npy | Neuropeptide Y | G-protein coupled receptor binding; neuropeptide signaling | (0.05) |
To confirm differential expression of selected upregulated genes from the microarray results shown in Table 1, western blot analysis was carried out on lysates collected from P19T1A2 cells treated with or without Dox (Figure 5A). All four of the protein products of selected genes found to be upregulated in the microarray—Gap43, Isl1, Synaptophysin, and Tau—were also upregulated in the presence of Dox. Each of these genes have previously been reported to play a role in neuronal differentiation and development (Mahalik et al., 1992; Jancsik et al., 1996; Jurata et al., 1996; Daly et al., 2000) and these results suggest that Ascl1 induction resulted in at least some transcriptional changes associated with neuronal differentiation.
Figure 5. P19T1A2 cells show characteristics of mature neurons.
A. Western blot analysis examining the protein expression of selected Ascl1-induced target genes identified in the microarrays. As predicted from the microarray results, expression of Gap43, Isl1, Synaptophysin (Syp), and Tau proteins increased in response to Dox-induced overexpression of Ascl1. B. P19T1A2 cells have the electrophysiological properties of neurons. Shown is a representative action potential-like waveform recorded from a P19T1A2 cell grown in the presence of Dox for six days. C, C′. P19T1A2 cells are polarized. Immunostaining for expression of the dendritic marker Map2 (red) and the axonal marker Neurofilament-L (NF-L, green). Nuclei were visualized with DAPI staining and appear blue. The boxed area in (C) is enlarged in panel (C′), with dendritic varicosities indicated by white arrowheads.
A hallmark of neurons is their ability to propagate electrical signals, so we carried out studies to determine whether P19T1A2 cells have the electrophysiological properties of neurons. Recordings were made from a total of eight cells that had been treated with Dox for six days and had an average resting membrane potential of -39.1 ± 0.6 mV. Five of the eight cells exhibited suprathreshold action potential-like waveforms. A representative action potential-like waveform is presented in Figure 5B. Another characteristic of differentiating neurons is their asymmetric development of processes into distinct axons and dendrites. The polarization of axons and dendrites underlies the ability of neurons to integrate and transmit information in the brain. These two types of processes differ from one another in morphology (Goslin and Banker, 1989; Craig and Banker, 1994), capacity for protein synthesis (Miyashiro et al., 1994), and in the molecular constituents of their cytoskeletons and plasma membranes (Barnes and Polleux, 2009). Using immunocytochemistry, we distinguished neurites expressing Map2, a marker of dendrites (Garner et al., 1988), from those that expressed the axonal marker neurofilament-L (NF-L; Szaro and Gainer, 1988; Figure 5C). The neurites that expressed Map2 had a shorter, tapered morphology characteristic of dendrites, and varicosities could be seen along the length of certain projections (Figure 5C′, arrowheads). The neurites that expressed NF-L were more elongated and slender, characteristic of axonal projections. These electrophysiological and immunocytochemical findings suggest that Dox-treated P19T1A2 cells share at least some aspects of neuronal development in common with primary neurons.
Identification of early changes in gene expression following Ascl1 induction
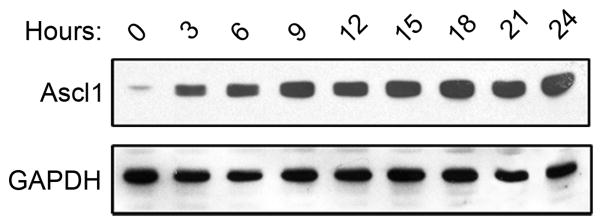
Having confirmed a subset of the microarray results and characterized neuronal differentiation of P19T1A2 cells, we sought to examine the earliest changes in gene expression following Ascl1 induction. P19T1A2 cells were cultured in the presence or absence of 0.5 μg/ml Dox for 0, 3, 6, 9, 12, 15, 18, 21, and 24 h. RNA and whole cell extracts were then isolated for microarray analysis, RT-PCR, and western blot analysis. Western blotting showed that Ascl1 protein was detectable as early as 3 h after treatment with Dox, and levels remained elevated throughout the 24 h time course (Figure 6).
Figure 6. Early Ascl1-expression in P19T1A2 cells.
Western blot analysis of P19T1A2 cells treated with Dox for the indicated hours. Ascl1 protein was induced as early as 3 h after the addition of Dox. Ascl1 expression became saturated at 9 h and remained elevated for the duration of the time course.
Microarray hybridization results indicated that the mRNA abundance for 28 known genes was induced greater than four-fold after 24 h of Dox treatment. Furthermore, the mRNA abundance for 14 other genes was found to be reduced by greater than four-fold over the same time course (data not shown). Selected genes that were induced or repressed strongly at early time points are shown in Table 2. After background subtraction, the mRNA for the Gadd45γ gene was found to show the greatest fold induction (33-fold) after 24 h of Dox treatment.
Table 2. Selected gene expression changes by microarray analysis at one day following Ascl1 induction.
| Gene Symbol | Gene Name(s) | Function/Process | Fold Change(s) |
|---|---|---|---|
| Gadd45γ | Growth arrest and DNA-damage-inducible 45 gamma | Protein binding; regulation of cell cycle | (33, 16) |
| Ankrd1 | Ankyrin repeat domain 1 | Transcription corepressor activity | (25, 16) |
| Unc45b | Unc-45 homolog B | Binding; cell differentiation | (19, 1.5) |
| Rgs16 | Regulator of G-protein signaling 16 | GTPase activator activity; GPCR signaling | (17) |
| Igfbp5 | Insulin-like growth factor binding protein 5 | Growth factor binding | (15, 13, 9) |
| Cdk5r1 | Cyclin-dependent kinase 5; regulatory subunit 1 (p35) | Kinase activity; axon guidance | (15) |
| Ngfr | Nerve growth factor receptor (p75NTR) | Receptor activity; apoptosis; axon guidance | (14, 1.2, 1.1) |
| Cdh3 | Cadherin 3 (P-cadherin) | Calcium ion binding; hemophilic cell adhesion | (13, 11, 5, 1.8) |
| Idb2 | Inhibitor of DNA binding 2 | Transcription regulator activity | (13) |
| Idb1 | Inhibitor of DNA binding 1 | Transcription regulator activity; BMP signaling | (12) |
| Nsg2 | Neuron specific gene family member 2 | Dopamine receptor binding; dopamine signaling | (11, 9, 1.0, 1.0) |
| Mfng | MFNG glucosaminyltransferase (manic fringe) | Glycosyl transferase activity; organismal development | (9.4, 4.0) |
| Nanog | Nanog homeobox | DNA binding; embryonic development | (0.21, 0.61) |
| Slc35d3 | Solute carrier family 35, member D3 (fringe-like 1) | Sugar:hydrogen symporter activity; transport | (0.21) |
| Glra2 | Glycine receptor, alpha 2 | GABA-A receptor activity; chloride channel activity | (0.19) |
| Tdgf1 | Teratocarcinoma-derived growth factor 1 (Cripto-1) | Growth factor activity; activation of MAPK activity | (0.19) |
| Eras | ES cell-expressed Ras (HRAS2) | GTP binding; small GTPase signal transduction | (0.17) |
| Trh | Thyrotropin releasing hormone | Hormone activity; hormone-mediated signaling | (0.16) |
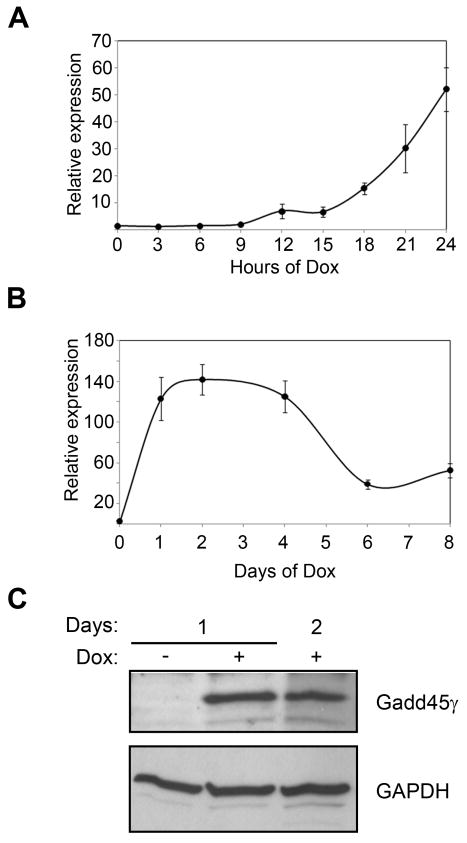
Validation of the microarray expression data for Gadd45γ was carried out using RT-PCR. Multiple oligonucleotide primer pairs were evaluated for detection of Gadd45γ mRNA (data not shown), and data generated from a representative set are shown. The induction patterns for Gadd45γ mRNA expression were in concordance with the microarray data: elevated expression levels of Gadd45γ mRNA were first detected 12 h after the addition of Dox, and expression continued to increase, reaching a 38-fold induction at 24 h (Figure 7A). Upon longer treatment with Dox, we continued to see elevated levels of Gadd45γ mRNA (Figure 7B). Using identical Dox exposure as for the microarray and RT-PCR experiments, western blot analysis was performed to confirm that the increase in Gadd45γ mRNA expression also resulted in increased levels of Gadd45γ protein (Figure 7C).
Figure 7. A novel Gadd45γ-immunoreactive protein unique to P19 cells is induced during neuronal differentiation.
A, B. RT-PCR analysis of early (A) and late (B) stages of Ascl1-induced neuronal differentiation showed that Gadd45γ mRNA expression increased in response to Ascl1, remained elevated through day four, and then declined. Results are shown as the mean ± s.d. normalized to GAPDH levels. C. Western blot analysis showing induction of Gadd45γ protein expression during Dox-induced differentiation of P19T1A2 cells.
Ascl1 regulates transcription of Gadd45γ and employs two proximal E-box consensus sites
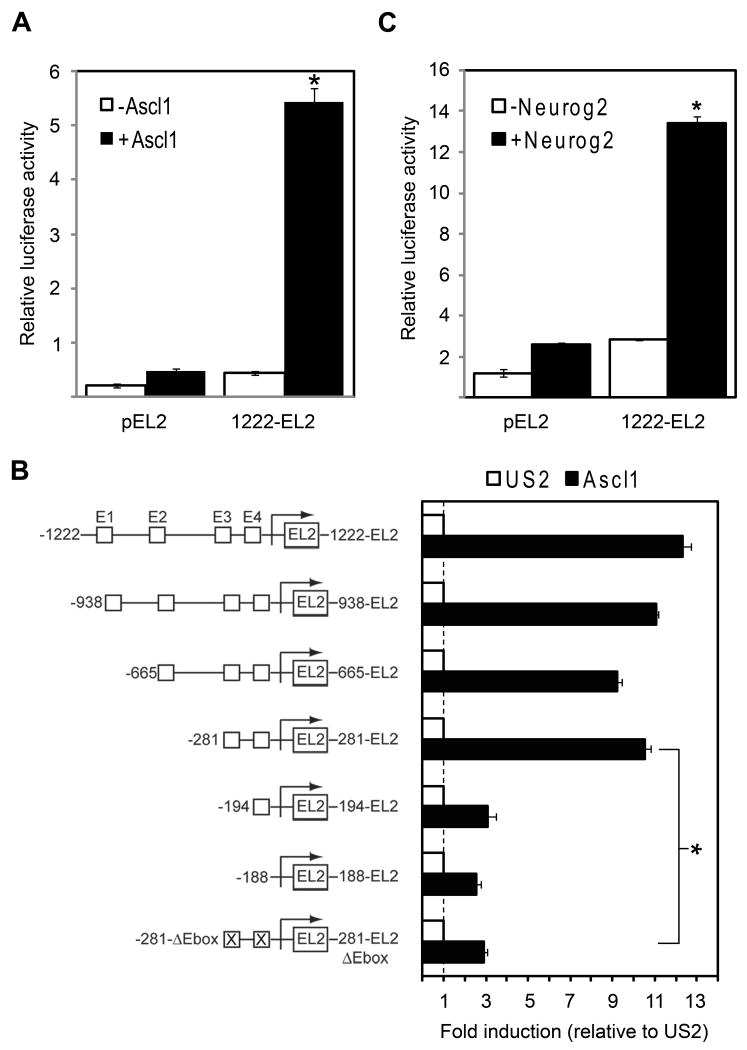
bHLH transcription factors such as Ascl1 usually function as transcriptional activators by binding to specific E-box motifs (CANNTG). As described in the Experimental Methods, we identified evolutionarily conserved sequences adjacent to the human and mouse Gadd45γ promoters. We further identified four E-box sequences clustered within these conserved sequences. The promoter region of the human Gadd45γ gene was subcloned into the promoter-less pEL2 reporter vector (1222-EL2), and its transcriptional properties were assayed in wild-type P19 cells via transient transfection in the presence or absence of Ascl1 (Figure 8A). A statistically significant, 12-fold increase in the activity of the 1222-EL2 reporter was observed in the presence of Ascl1, consistent with our microarray and RT-PCR results showing induction of Gadd45γ transcription by Ascl1 (see Table 2 and Figure 7). All of the 5′ Gadd45γ truncated constructs except 194- and 188-EL2 were able to drive transcription of the EL2 reporter in P19 cells, suggesting that the 281 bp fragment contains the core promoter elements sufficient to drive transcription in response to Ascl1 (Figure 8B). As described in the Experimental Methods, we introduced 4 bp substitution mutations in the two E-box consensus sites designated E3 and E4 and tested their effect on promoter activity. Figure 8B also shows the relative reduction in promoter activity of the mutated (281-EL2ΔEbox) versus wild-type (281-EL2) reporter. Mutations in the two E-box sites significantly reduced promoter activity by 4.2-fold, suggesting that the ability of Ascl1 to activate transcription of Gadd45γ was dependent on the presence of these two E-boxes in the promoter.
Figure 8. Direct transcriptional regulation of Gadd45γ by Ascl1.
A. Co-transfection of the Gadd45γ reporter with Ascl1 yielded a relative luciferase activity of 5.4 as compared to 0.46 in the absence of Ascl1 (a 12-fold increase). Results are expressed as mean relative luciferase activity, with error bars denoting standard deviation. *p < 0.01. B. Transcriptional activity of 5′ Gadd45γ deletion reporters in response to Ascl1. Schematics of pEL2 reporter constructs used are shown on the y-axis. Deletion analyses localized the necessary Ascl1 regulatory element to within 281 bp upstream of the transcriptional start site, and mutational analysis showed that augmentation of Gadd45γ transcription was dependent on the presence of two proximal E-boxes. * p < 0.01 as compared to the wild-type 281-EL2 reporter. C. Co-transfection of the Gadd45γ reporter with Neurog2 yielded a relative luciferase activity of 13 as compared to 2.6 in the absence of Neurog2 (a 5.2-fold increase). Results are expressed as mean relative luciferase activity, with error bars denoting standard deviation. *p < 0.01.
Recent research has shown cooperative activity between Ascl1 and the POU proteins Brn1 and Brn2 in mediating expression of certain genes critical for neurogenesis (Castro et al., 2006). However, the octameric motif recognized by the Brn transcription factors was not seen in the regions surrounding the essential E-boxes in the Gadd45γ promoter. Furthermore, VISTA analysis did not show any other highly conserved sequences in the Gadd45γ promoter region (data not shown). Therefore, the reduction of reporter activity driven by the Gadd45γ promoter was specifically due to loss of Ascl1 interaction with the core promoter region, and not due to loss of Ascl1 interaction with other DNA-binding cofactors such as the Brns.
Other bHLH factors are known to function together to regulate development of the nervous system (Bertrand et al., 2002). Ascl1 and the Neurogenin family constitute the main proneural proteins in mammals, and research has shown that they can cooperatively regulate neural progenitor cell cycle exit, the specification of neuronal subtype identities, and neuronal migration (Bertrand et al., 2002; Hand et al., 2005; Ge et al., 2006). Recently, the integrated activity of Ascl1 and Neurogenin-2 (Neurog2) with specific E-boxes was shown to temporally regulate Dll3 levels during neural tube development (Henke et al., 2009). To determine whether Neurog2 can also regulate the Gadd45γ promoter, the full-length 1222-EL2 reporter was transiently transfected into P19 cells in the presence or absence of Neurog2 (Figure 8C). A significant, 5.2-fold increase was observed in the presence of Neurog2, consistent with preliminary microarray data showing induction of Gadd45γ transcription by Neurog2, but overall to a lesser extent than the induction by Ascl1 (data not shown).
Ascl1 binds directly to the Gadd45γ promoter
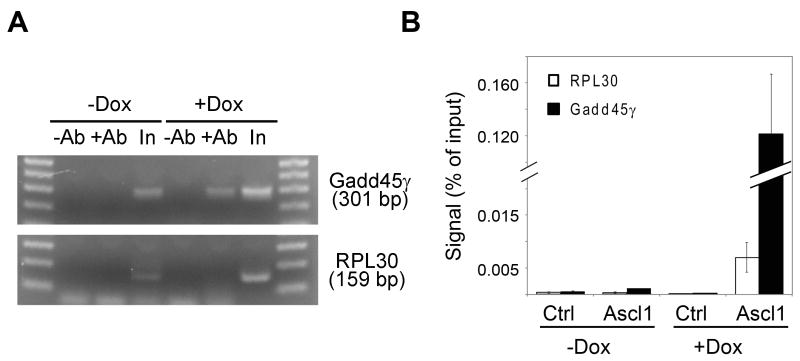
To determine whether Ascl1 could bind directly to the Gadd45γ promoter in P19 cells, we performed chromatin immunoprecipitation (ChIP) experiments. Chromatin was immunoprecipitated with an antibody specific to Ascl1 from formaldehyde cross-linked P19T1A2 cells treated with or without Dox for 24 h. To determine whether Ascl1 localized to the Gadd45γ promoter, quantitative RT-PCR amplification was performed using primers encompassing the proximal E-box sequences (E3/E4). Figure 9A shows representative ChIP-PCR samples that were stopped in the linear amplification range, run on an agarose gel, and visualized with ethidium bromide. Chromatin immunoprecipitated with Ascl1 antibody from P19T1A2 cells treated with Dox showed significant enrichment (10-fold) for the Gadd45γ promoter sequence containing E-boxes E3 and E4. Negative controls with primers specific to the RPL30 gene had no significant enrichment. In Figure 9B, the ChIP-PCR amplification products were quantified and normalized to the input of each sample. Taken together, these results demonstrate that Ascl1 directly binds to the Gadd45γ promoter in differentiating P19T1A2 cells.
Figure 9. Ascl1 occupies the regulatory regions containing E-boxes E3 and E4 of Gadd45γ in P19T1A2 cells.
A. P19T1A2 cells treated with or without Dox for 24 h were subjected to ChIP assays using anti-Ascl1 antibody (+Ab) or control IgG (-Ab) followed by real-time PCR assays to detect the Gadd45γ and RPL30 promoter DNAs. Representative ChIP-PCR analyses were stopped in the linear amplification range and run on an agarose gel for visualization with ethidium bromide. Input (In) samples were loaded as a control. B. ChIP-PCR analysis of the binding of Ascl1 to Gadd45γ enhancers in P19T1A2 cells using primers specific to Gadd45γ (black bars) or a control gene (RPL30; white bars). Data are presented as the mean percentage of input ± s.d. of the results from three independent experiments.
Gadd45γ is sufficient to induce a neuronal-like phenotype in P19 cells
In order to more closely examine the transcriptional events initiated by expression of Gadd45γ, a stable P19 cell line (P19T3GIE2) in which expression of both Gadd45γ and EGFP was under control of the TRE promoter was generated (see Experimental Methods). One day after induction of Gadd45γ with Dox, no TuJ1-immunoreactive cells were seen (Figure 10A). By eight days, however, a fraction of P19T3GIE2 cells adopted a neuronal morphology and expressed TuJ1 (Figure 10B). Preliminary microarray hybridization analyses showed a number of commonly induced or repressed genes between P19T1A2 and P19T3GIE2 cells. The identities, associated functions, and mRNA fold changes of select genes are provided in Table 3.
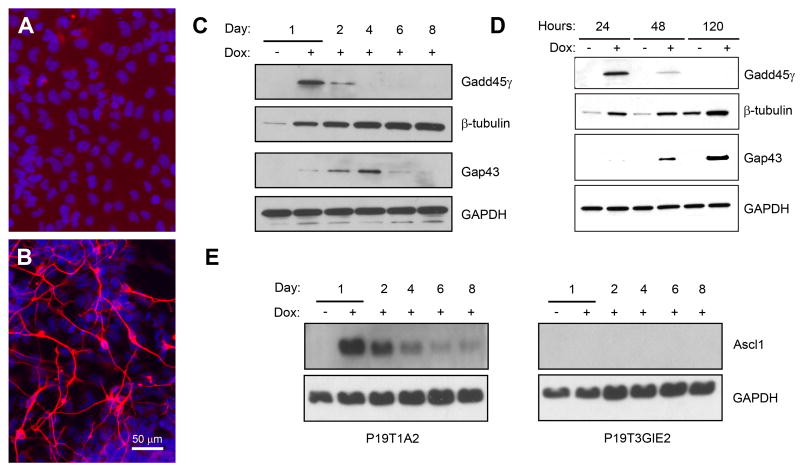
Figure 10. Overexpression of Gadd45γ is sufficient to induce a neuronal-like phenotype.
A-B. TuJ1 (red) staining of P19T3GIE2 cells one day (A) and eight days (B) after treatment with Dox. Nuclei were visualized with DAPI and appear blue. Eight days after treatment with Dox, P19T3GIE2 cells adopted a neuronal morphology and expressed TuJ1. Scale bar = 50 μm. C. Western blotting for protein expression in P19T3GIE2 cells treated with Dox for the indicated days. Gadd45γ was induced one day post-Dox, and then declined. P19T3GIE2 cells also showed induction of proteins characteristic of a P19T1A2 induction, e.g. β-III-tubulin and Gap43. D. Western blotting using antibodies against β-III-tubulin and Gap43 showed that overexpression of Gadd45γ was sufficient to induce expression of neuronal protein markers in a Dox-dependent manner. E. Western blot analysis for Ascl1 expression in P19T1A2 and P19T3GIE2 cells. Ascl1 protein expression is strongly induced in P19T1A2 cells one day after treatment with Dox and then declines. No Ascl1 expression was detected in P19T3GIE2 cells at any time point.
Table 3. Selected gene expression changes by microarray analysis at eight days following Gadd45γ induction.
| Gene Symbol | Gene Name(s) | Function/Process | Fold Change(s) |
|---|---|---|---|
| Igfbp5 | Insulin-like growth factor binding protein 5 | Growth factor binding | (20, 20, 15) |
| Dner | Delta/notch-like EGF-related receptor | Notch binding | (9) |
| Map2 | Microtubule-associated protein 2 | Cytoskeletal regulatory protein binding | (6) |
| Slc17A6 | Solute carrier family 17; Vglut2 | L-glutamate transmembrane transporter activity | (15, 13) |
| Svop | SV2 related protein | Ion transmembrane transporter activity | (5) |
| Syt4 | Synaptotagmin IV | Calcium ion binding; neurotransmitter secretion | (8) |
| Pou5f1 | POU domain, class 5, transcription factor 1 | DNA binding; cell fate commitment | (0.18, 0.17) |
| FGF5 | Fibroblast growth factor 5 | Growth factor activity; cell proliferation | (0.16) |
| Lefty | Left right determination factor 1 | Growth factor activity; anterior/posterior axis specification | (0.06) |
| Npy | Neuropeptide Y | G-protein coupled receptor binding; neuropeptide signaling | (0.06) |
Western blot analysis at various times following Dox treatment confirmed that P19T3GIE2 cells showed induction of some proteins that are characteristic of a P19T1A2 differentiation program such as β-III-tubulin and Gap43 (Figure 10C). β-III-tubulin protein was detected as early as one day post-Dox treatment, with a gradual increase up to eight days. Gap43 protein was detected two days post-Dox treatment, increased at four days, and then declined. Map2 protein expression was not detected at any time point by western blot or immunocytochemistry following Dox treatment of P19T3GIE2 cells (data not shown). Therefore, it appears that while Gadd45γ is sufficient to induce some Ascl1 induced proteins (such as β-III-tubulin and Gap43), it is not sufficient to induce others (such as Map2).
In order to validate the induction of β-III-tubulin and Gap43 by Gadd45γ as being Dox-dependent, we conducted western blot analysis on lysates from cells treated with and without Dox at 24, 48, and 120 h (Figure 10D). A clear induction of β-III-tubulin protein was seen in P19T3GIE2 cells treated with Dox at all time points shown. Western blot analysis also showed an increase in Gap43 protein at 48 and 120 h. The clear difference in β-III-tubulin expression at earlier time points between cells treated with or without Dox combined with the induction of Gap43 only in Dox-treated cells strongly demonstrates that overexpression of Gadd45γ is sufficient to induce a subset of Ascl1 transcriptional responses in P19 cells. Microarray hybridization analyses showed that Gadd45γ induction did not induce expression of bHLH proteins including Ascl1, Neurog2, and NeuroD2 (data not shown). Western blot analysis confirmed that Ascl1 protein is not induced in P19T3GIE2 cells (Figure 10E). Together, these data show that Ascl1 is not required for the Gadd45γ-induced transcriptional responses of β-III-tubulin or Gap43.
Discussion
Despite the importance of neurogenic bHLH families during neuronal development, knowledge of their physiological target genes is still incomplete. Ascl1 is one of the earliest markers expressed in neural progenitor cells and is essential for their survival and differentiation (Parras et al., 2004). Recently, Ascl1 was found to be the only gene within a pool of 19 candidate genes that was sufficient to induce neuron-like cells in mouse fibroblasts (Vierbuchen et al., 2010). Therefore, delineation of the gene regulatory networks controlled by Ascl1 is critical to understanding the transcriptional interactions that control neuronal differentiation. A major challenge to elucidating the Ascl1-induced genetic cascades is the cellular complexity of the developing embryo, as well as the limited number of defined cells that can be obtained from each embryo. These shortcomings may be circumvented by the use of in vitro models, such as the P19T1A2 cells characterized here. The P19T1A2 cells demonstrated tight regulation of Ascl1, and varying the levels of Ascl1 expression resulted in varying extents of neuronal differentiation and neurons that were electrically excitable (see Figure 5). The P19T1A2 cells can therefore provide a continuous source for generating a large number of stage-specific cells, which facilitates many types of analyses, including large-scale genomic profiling via microarray analysis.
Achieving stable expression of transfected genes in P19 cells has historically been more problematic than other continuous cell lines (McBurney, 1993). Clonal populations of transformed P19 cells often segregate into non-expressing variants that can rapidly become predominant in the population. At least partially, the successful generation of the P19T1A2 cell line was due to use of the human ubiquitin C promoter, first exon, first intron and partial second exon, which resulted in high, sustainable levels of rtTA expression in the P19 cells. This high efficiency of the ubiquitin C promoter in P19 cells has been reported previously (Yu et al., 2005).
To gain insight into the role of Ascl1 in neuronal development, we characterized downstream transcriptional targets of Ascl1. Using microarray hybridization assays of P19T1A2 cells induced to differentiate in the presence of Dox, we identified several genes showing differential expression that are also known to be important for neurite outgrowth, axon guidance, and differentiation. For example, FGF5 is frequently expressed in embryonic tissues and has recently been described as an embryonic stem cell marker (Pelton et al., 2002). We observed a 9-fold decrease in its mRNA expression after eight days of neuronal differentiation (see Table 1). In concordance with this data, studies have shown that FGF5 mRNA expression is inhibited during retinoic acid-induced neuronal differentiation (Martinez-Ceballos et al., 2005), and its expression also decreases upon specification of embryonic stem cells to a neuroectodermal fate (Shimozaki et al., 2003). These induced neuronal cells displayed functional neuronal properties such as the generation of trains of action potentials and synapse formation, as well as polarization into dendritic and axonal domains. The present work also identified a number of genes that were not previously known candidates for regulating neuronal differentiation. For example, microarray hybridization showed that Gadd45γ was the earliest and most highly induced gene, a finding confirmed by RT-PCR and western blot analysis.
Differentiation of neuronal precursors is characterized by a loss of multipotency and cell-cycle exit. Previous studies have shown that simultaneous with the differentiation program induced by proneural bHLH proteins, an anti-proliferative response is also induced through the upregulation of cell cycle inhibitors such as the cyclin kinase inhibitor p27kip1 (Farah et al., 2000). Several studies have shown that the Gadd45 proteins also regulate the cell cycle via interactions with PCNA (Smith et al., 1994), cyclin-dependent kinase inhibitor p21 (Yang et al., 2000), and Cdc2 to inhibit Cdc2-cyclin B1 kinase activity (Zhan et al., 1999). In addition, Gadd45 proteins activate the p38/Jun N-terminal kinase pathway by binding to MTK1/MEKK4 in response to environmental stress (Takekawa and Saito, 1998). Induction of Gadd45 genes in cell culture was shown to stop the cell cycle in G1 phase (Zhang et al., 2001), which is compatible with cell cycle exit—a requirement for terminal neuronal differentiation. Gadd45γ was identified in the medaka Oryzias latipes as a gene differentially expressed in the regions in which cells stop dividing and begin differentiating, e.g. the optic tectum, and the hypothalamic and telencephalic ventricles (Candal et al., 2004). It is therefore plausible that the upregulation of Gadd45γ observed in response to Ascl1 is important for the cell cycle withdrawal that precedes neuronal differentiation.
Novel roles for Gadd45 are also becoming apparent: a recent study identified Gadd45 as one factor in a system of proteins involved in the demethylation process in zebrafish embryos (Rai et al., 2008), and Gadd45β was found to be required for activity-induced DNA demethylation of specific promoters and expression of corresponding genes critical for adult neurogenesis (Ma et al., 2009). Since upregulation of Gadd45 proteins affects cell cycle regulation, cell survival, and cell death—all of which are important processes during neuronal development—and because Gadd45γ is one of the earliest and most highly expressed genes in our model system of neuronal differentiation, it will be important in future studies to determine which of these multiple roles Gadd45γ fulfills in the gene regulatory network that guides Ascl1-induced neuronal differentiation.
In preliminary studies, we were unable to determine whether Gadd45γ was required for Ascl1-induced differentiation, because five shRNA constructs targeted against the mouse Gadd45γ gene were ineffective in sufficiently reducing Gadd45γ mRNA or protein levels in P19T1A2 cells. Despite this, our findings that Ascl1 overexpression drives P19 cells towards exit from the cell cycle and generation of neurons in vitro, and that Gadd45γ is strongly induced soon after Ascl1 induction suggests that Gadd45γ couples cell cycle exit to neuronal differentiation.
Research has shown that regulation of histone acetylation/deacetylation levels is essential for murine Gadd45γ promoter control, and that functional Oct and NF-Y elements are essential for basal expression of the promoter (Campanero et al., 2008). Gadd45γ has also previously been implicated as a transcriptional target of bHLH proteins. In Xenopus, injection of Ngnr1 or NeuroD promoted ectopic expression of Gadd45γ (de la Calle-Mustienes et al., 2002). Furthermore, Gadd45γ was identified as a direct NeuroD responsive target gene, with conserved induction in mammalian cells (Seo et al., 2007). Microarray studies from gain- and loss-of-function analyses in developing mouse dorsal or ventral telencephalon also identified Gadd45γ as an Ascl1 target gene, with expression predominantly in the subventricular zone (Gohlke et al., 2008). However, these previous studies did not identify promoter elements required for Gadd45γ transcriptional regulation by Ascl1 and did not characterize the Gadd45γ protein.
Our results provide the first evidence for a direct regulation of the Gadd45γ gene by Ascl1. We demonstrated the importance of a 281 bp region of the Gadd45γ promoter for Ascl1 induction: when E-boxes within this region were destroyed, the promoter displayed almost complete loss of Ascl1-induced activity (see Figure 8). Furthermore, ChIP-PCR analysis showed that Ascl1 binds directly to the Gadd45γ promoter in differentiating P19T1A2 cells (see Figure 9). The overlap between the Ascl1 and Gadd45γ gene regulatory networks suggests that Gadd45γ acts downstream of Ascl1 in vitro (see Figure 10 and Table 3). However, we have also identified genes belonging to the Ascl1 gene regulatory network, such as Map2 and Synaptophysin, whose expression appears not to be affected by over-expression of Gadd45γ (data not shown). These data suggest that Ascl1 requires the induction of other genes in addition to Gadd45γ in order to generate a more complete neuronal differentiation program. Recent research has shown that Ascl1 alone is sufficient to induce some neuronal traits in mouse fibroblasts, but additional factors such as Brn2 and Myt1l are necessary to facilitate neuronal conversion and maturation (Vierbuchen et al., 2010). In our microarray hybridization assays of P19T1A2 cells treated with Dox, we also see an increase of Myt1l expression after Ascl1 induction, supporting the conclusion that while Ascl1 is sufficient to induce immature neuronal features, expression of other downstream factors are necessary to generate mature neurons with high efficiency.
Neuronal differentiation in vivo is the result of extrinsic cues such as retinoic acid, bone-morphogenic antagonists, and cell surface molecules activating and altering intrinsic genetic programs within neural stem and progenitor cells. The bHLH proteins, such as Ascl1, function at critical points in these genetic programs to generate fully differentiated neurons at the proper developmental time and anatomical position within the embryo (Rodriguez et al., 1990; Guillemot et al., 1993; Horton et al., 1999). The P19T1A2 cells in which Ascl1 is tightly regulated by Dox will allow for more detailed elucidation of Ascl1 regulated genetic programs. While microarray hybridization assays are beneficial for discerning gene regulatory networks of neuronal differentiation, the importance of determining global changes in protein expression could also significantly enhance the fidelity of network modeling. Protein translation and stability are often regulated separately from mRNA, and as such there is often a lack of correlation between changes in protein levels and changes in mRNA levels. Furthermore, post-translational modifications such as phosphorylation, proteolysis, and ubiquitination can drastically alter protein function but are outside the scope of microarray hybridization studies. The ease with which the P19T1A2 cells can be grown, and their reliability to yield a high percentage of cells expressing neuronal proteins makes this cell line a robust system for proteomic analysis as well as for high-throughput chemical and RNAi screening. In the long term, such studies may help to guide the development of differentiation strategies for human embryonic stem cells in the treatment of human neurodegenerative diseases.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the helpful discussions of Dr. Fan Meng and the critical review of the manuscript by Dr. Stephen Fisher. This work utilized Proteomics Core(s) of the Michigan Diabetes Research and Training Center, funded by DK020572 from the National Institute of Diabetes and Digestive and Kidney Diseases. This work was supported by NIH/NINDS R01 NS051472 (MDU).
Abbreviations used
- bHLH
basic helix-loop-helix
- ChIP
chromatin immunoprecipitation
- CNS
central nervous system
- CS
calf serum
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- Dox
doxycycline
- EC
embryonic carcinoma
- EGFP
enhanced green fluorescent protein
- FBS
fetal bovine serum
- Gadd45γ
growth-arrest and DNA-damage inducible protein 45 gamma
- Map2
microtubule-associated protein 2
- Ascl1
mammalian achaete-scute homolog 1
- MEMα
minimal essential medium alpha
- NF-L
neurofilament-L
- Neurog2
neurogenin-2
- P19T1A2
Ascl1-inducible P19 cell line
- P19T3GIE2
Gadd45γ-inducible P19 cell line
- PBS
phosphate buffered saline
- DPBS
Dulbecco's PBS
- PNS
peripheral nervous system
- RPL30
ribosomal protein L30
- RT-PCR
real-time quantitative PCR
- rtTA
reverse tetracycline responsive transactivator
- TuJ1
neuron-specific class III β-tubulin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bain G, Ray WJ, Yao M, Gottlieb DI. Retinoic acid promotes neural and represses mesodermal gene expression in mouse embryonic stem cells in culture. Biochem Biophys Res Commun. 1996;223(3):691–694. doi: 10.1006/bbrc.1996.0957. [DOI] [PubMed] [Google Scholar]
- Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Thieke K, Maier A, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35(5):567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Campanero MR, Herrero A, Calvo V. The histone deacetylase inhibitor trichostatin A induces GADD45γ expression via Oct and NF-Y binding sites. Oncogene. 2008;27:1263–1272. doi: 10.1038/sj.onc.1210735. [DOI] [PubMed] [Google Scholar]
- Candal E, Thermes V, Joly JS, Bourrat F. Medaka as a model system for the characterization of cell cycle regulators: a functional analysis of Ol-Gadd45γ during early embryogenesis. Mech Devel. 2004;121:945–958. doi: 10.1016/j.mod.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Carbonetto S, Gruver MMS, Turner DC. Nerve fiber growth in culture on fibronectin, collagen, and glycosaminoglycan substrates. J Neurosci. 1983;3:2324–2335. doi: 10.1523/JNEUROSCI.03-11-02324.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Göttgens B, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11(6):831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Collins SP, Uhler MD. Cyclic AMP- and cyclic GMP-dependent protein kinases differ in their regulation of cyclic AMP response element-dependent gene transcription. J Biol Chem. 1999;274(13):8391–8404. doi: 10.1074/jbc.274.13.8391. [DOI] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Daly C, Sugimori M, Moreira JE, Ziff EB, Llinas R. Synaptophysin regulates clathrin-independent endocytosis of synaptic vesicles. Proc Natl Acad Sci USA. 2000;97(11):6120–6125. doi: 10.1073/pnas.97.11.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E, Glavic A, Modolell J, Gómez-Skarmeta JL. Xiro homeoproteins coordinate cell cycle exit and primary neuron formation by upregulating neuronal-fate repressors and downregulating the cell-cycle inhibitor XGadd45-gamma. Mech Dev. 2002;119(1):69–80. doi: 10.1016/s0925-4773(02)00296-4. [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Garner CC, Tucker RP, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI, Martinowich K, et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci USA. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke JM, Armant O, Parham FM, et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González F, Romani S, Cubas P, Modolell J, Campuzano S. Molecular analysis of the asense gene, a member of the achaete-scute complex of Drosophila melanogaster, and its novel role in optic lobe development. EMBO J. 1989;8(12):3553–3562. doi: 10.1002/j.1460-2075.1989.tb08527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslin K, Banker G. Experimental observations on the development of polarity by hippocampal neurons in culture. J Cell Biol. 1989;108:1507–1516. doi: 10.1083/jcb.108.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75(3):463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Henke RM, Meredith DM, Borromeo MD, Savage TK, Johnson JE. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol. 2009;328(2):549–540. doi: 10.1016/j.ydbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor ASCL1. Mol Cell Neurosci. 1999;14(4-5):355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- Jancsik V, Filliol D, Rendon A. Tau proteins bind to kinesin and modulate its activation by microtubules. Neurobiology (Bp) 1996;4:417–429. [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Anderson DJ. Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature. 1990;346(6287):858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Zimmerman K, Saito T, Anderson DJ. Induction and repression of mammalian achaete-scute homologue (MASH) gene expression during neuronal differentiation of P19 embryonal carcinoma cells. Development. 1992;114(1):75–87. doi: 10.1242/dev.114.1.75. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA. 1996;93(21):11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochter A, Schachner M. Tenascin and extracellular matrix glycoproteins: from promotion to polarization of neurite growth. in vitro J Neurosci. 1993;13:3986–4000. doi: 10.1523/JNEUROSCI.13-09-03986.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalik TJ, Carrier A, Owens GP, Clayton G. The expression of GAP43 mRNA during late embryonic and early postnatal development of the CNS of the rat: an in situ hybridisation study. Brain Res Dev Brain Res. 1992;67:75–83. doi: 10.1016/0165-3806(92)90027-t. [DOI] [PubMed] [Google Scholar]
- Martinez-Ceballos E, Chambon P, Gudas LJ. Differences in gene expression between wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitor factor (LIF) removal. J Biol Chem. 2005;280(16):16484–16498. doi: 10.1074/jbc.M414397200. [DOI] [PubMed] [Google Scholar]
- McBurney MW, Jones-Villeneuve EM, Edwards MK, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299(5879):165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- McBurney MW. P19 embryonal carcinoma cells. Int J Dev Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neuritis: implications for neuronal functioning. Proc Natl Acad Sci USA. 1994;91(23):10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacherník J, Bryja V, Esner M, Kubala L, Dvorák P, Hampl A. Neural differentiation of pluripotent mouse embryonal carcinoma cells by retinoic acid: inhibitory effect of serum. Physiol Res. 2005;54(1):115–122. doi: 10.33549/physiolres.930526. [DOI] [PubMed] [Google Scholar]
- Parras CM, Galli R, Britz O, Soares S, Galichet C, Battiste J, Johnson JE, Nakafuku M, Vescovi A, Guillemot F. Ascl1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J. 2004;23(22):4495–4505. doi: 10.1038/sj.emboj.7600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton TA, Sharma S, Schulz TC, Rathjen J, Rathjen PD. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–339. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- Powell LM, Zur Lage PI, Prentice DR, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol. 2004;24(21):9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J, Raymon HK, Gage FH. Generation and culturing of precursor cells and neuroblasts from embryonic and adult central nervous system. Methods Enzymol. 1995;254:20–37. doi: 10.1016/0076-6879(95)54004-0. [DOI] [PubMed] [Google Scholar]
- Rodríguez I, Hernández R, Modolell J, Ruiz-Gómez M. Competence to develop sensory organ is temporally and spatially regulated in Drosophila epidermal primordial. EMBO J. 1990;9(11):3583–3592. doi: 10.1002/j.1460-2075.1990.tb07569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Letourneau PC, Palm SL, McCarthy J, Furcht LT. Neurite extension by peripheral and central nervous system neurons in response to substratum-bound fibronectin and laminin. Dev Biol. 1983;98:212–220. doi: 10.1016/0012-1606(83)90350-0. [DOI] [PubMed] [Google Scholar]
- Romani S, Campuzano S, Macagno ER, Modolell J. Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes Dev. 1989;3(7):997–1007. doi: 10.1101/gad.3.7.997. [DOI] [PubMed] [Google Scholar]
- Schrag JD, Jiralerspong S, Banville M, Jaramillo ML, O'Connor-McCourt MD. The crystal structure and dimerization interface of GADD45γ. Proc Natl Acad Sci USA. 2008;105(18):6566–6571. doi: 10.1073/pnas.0800086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozaki K, Nakashima K, Niwa H, Taga T. Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development. 2003;130:2505–2512. doi: 10.1242/dev.00476. [DOI] [PubMed] [Google Scholar]
- Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O'Connor PM, Fornace AJ., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- Szaro BG, Gainer H. Identities, antigenic determinants, and topographic distributions of neurofilament proteins in the nervous systems of adult frogs and tadpoles of Xenopus laevis. J Comp Neurol. 1988;273:344–358. doi: 10.1002/cne.902730306. [DOI] [PubMed] [Google Scholar]
- Takekawa M, Saito H. A Family of Stress-Inducible GADD45-like Proteins Mediate Activation of the Stress-Responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95(4):521–553. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Zhan Q, Coursen JD, et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Manicone A, Coursen JD, Linke SP, Nagashima M, Forgues M, Wang XW. Identification of a functional domain in a GADD45-mediated G2/M checkpoint. J Biol Chem. 2000;275:36892–36898. doi: 10.1074/jbc.M005319200. [DOI] [PubMed] [Google Scholar]
- Yi SH, Jo AY, Park CH, et al. Ascl1 and Neurogenin2 enchance survival and differentiation of neural precursor cells after transplantation to rat brains via deistinct modes of action. Mol Therapy. 2008;16(11):1873–1882. doi: 10.1038/mt.2008.189. [DOI] [PubMed] [Google Scholar]
- Yu JY, Wang TW, Vojtek AB, Parent JM, Turner DL. Use of short hairpin RNA expression vectors to study mammalian neural development. Methods Enzymol. 2005;392:186–199. doi: 10.1016/S0076-6879(04)92011-3. [DOI] [PubMed] [Google Scholar]
- Zhan Q, Antinore MJ, Wang XW, Carrier F, Smith ML, Harris CC, Fornace AJ., Jr Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892–2900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hoffman B, Liebermann DA. Ectopic expression of MyD118/Gadd45/CR6 (Gadd45beta/alpha/gamma) sensitizes neoplastic cells to genotoxic stress-induced apoptosis. Int J Oncol. 2001;18(4):749–757. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.