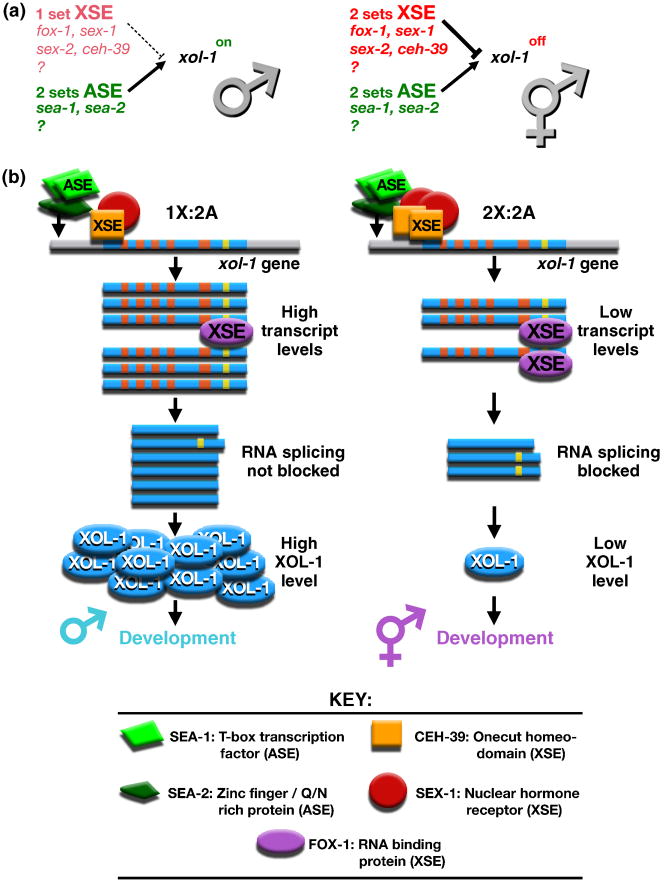
Figure 1. Primary sex determination: the X:A signal model.
(a) In C. elegans, sex is determined by the number of X chromosomes relative to the ploidy, the number of sets of autosomes. This X:A signal is composed of X signal elements (XSEs) that activate the hermaphrodite program of development and Autosomal signal elements (ASEs) that oppose the XSEs to promote male development. The direct target of the X:A signal is the sex determination switch gene xol-1, which controls both the choice of sexual fate and the level of X-linked gene expression achieved through the process of dosage compensation.
(b) X and autosomal signals antagonize each other directly at xol-1 to determine C. elegans sex. Direct rivalry at the xol-1 promoter between XSE transcriptional repressors (the ONECUT homeodomain protein CEH-39 and the nuclear hormone receptor SEX-1) and ASE transcriptional activators (the T-box transcription factor SEA-1 and the zinc finger protein SEA-2) leads to high xol-1 transcript levels in XO embryos and low levels in XX embryos. FOX-1, an RNA binding protein that acts as an XSE, then enhances the fidelity of signaling process by creating an inactive xol-1 mRNA splice variant in a dose-dependent manner. High XOL-1 protein levels in XO animals cause male development, and low XOL-1 levels in XX animals cause hermaphrodite development, including loading of the DCC onto X. Decreasing XSE dose causes XX-specific lethality, while increasing it causes XO-specific lethality. The reciprocal is true for ASEs. Increasing ASE dose causes XX-specific lethality; decreasing it causes XO-specific lethality. (xol-1 gene: promoter, gray; exons, blue; introns, orange and yellow).