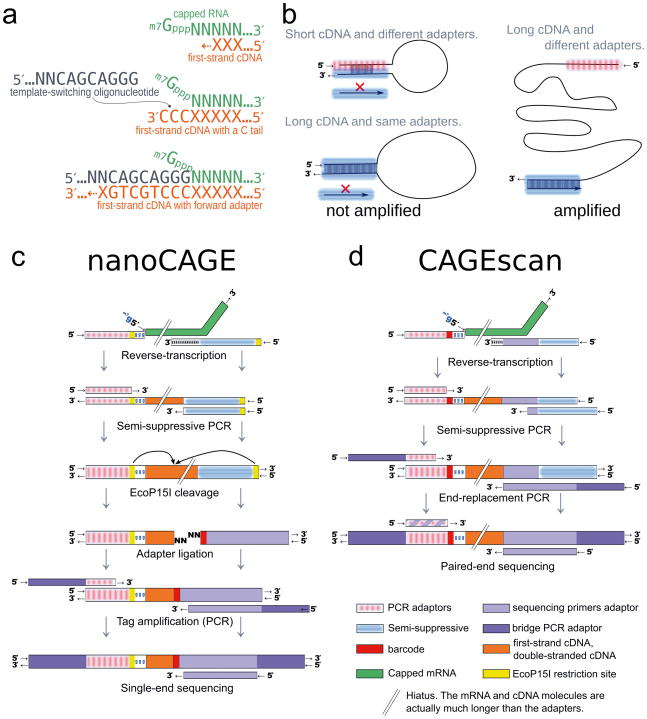
Figure 1. Experimental outline of the nanoCAGE and CAGEscan protocols.
(a) nanoCAGE captures the 5′ ends of molecules by template switching. When polymerizing the cDNA of a capped mRNA, the reverse transcriptase adds extra cytosines that are complementary to the cap. Thus each 5′ full-length cDNAs is extended upon hybridization of the riboguanosine-tailed “template-switching” oligonucleotides to these extra cytosines. (b) In the semi-suppressive PCR, the short templates fold intramolecularly and prevent the binding of primers which precludes amplification; longer molecules are less likely to fold and are thus amplified. Templates derived from reaction artifacts form stable homo-duplexes also precluding amplification. (c) Preparation of nanoCAGE tags. After template-switching, semi-suppressive PCR and EcoP151 cleavage, 25 bp are ligated to oligonucleotide adapters that contain a sequence identifier (red box). After PCR amplification, the nanoCAGE tags are subjected to sequencing by synthesis. (d) Preparation of 5′-full-length cDNA libraries for paired-end sequencing with the CAGEscan protocol. Capped mRNAs capture is similar to a. The ends of the amplified cDNA constructs are replaced by PCR with adapters for sequencing in the Illumina Genome Analyzer, that produces paired-end reads from single cDNAs.