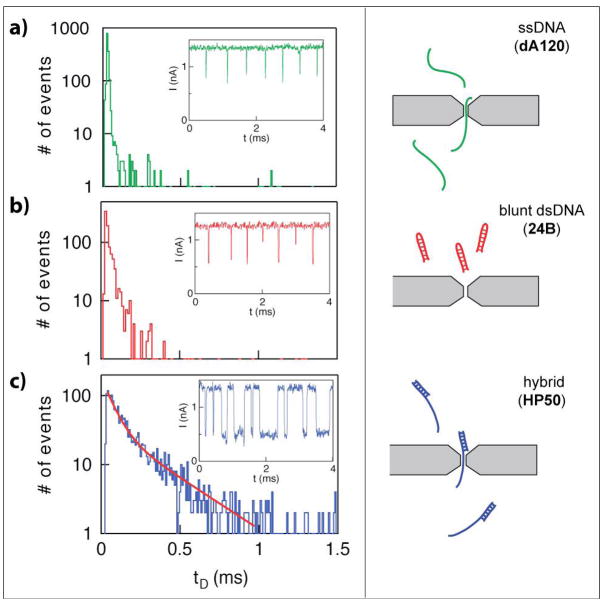
Figure 2.
Dwell-time distributions for three DNA samples through 2 nm pores. a) a ssDNA molecule consisting of 120-mer poly-deoxyadenine molecule. b) blunt dsDNA, consisting of a 24-bp duplex region and a 6-base loop. c) a hybrid single-strand/duplex molecule, consisting of a 100-mer strand hybridized to a fully-complementary 50-mer at one end. While the dwell-times for ssDNA and blunt dsDNA are short (< 21 μs), noticeably longer dwell-times are observed for the hybrid molecule. The complete dwell-time distribution for the hybrid molecule can be modeled as a bi-exponential function with decay timescales t0 = 40 μs and t1 = 240 μs, corresponding to a collision and translocation/unzipping process, respectively. Typical events for the three samples are shown as insets to the dwell-time distributions.