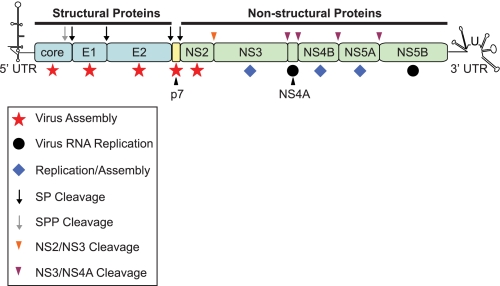
FIGURE 1.
Schematic representation of the HCV genome. The single open reading frame encodes 10 viral proteins that are divided into the structural (core, E1, and E2; shown in blue) and non-structural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B; shown in green) regions. The small peptide p7 is currently unassigned into either category (shown in yellow). The open reading frame is flanked by untranslated regions (UTR) at the 5′ and 3′ termini that are involved in both replication and translation. Following translation, the viral polyprotein is processed by two cellular protease activities, signal peptidase (SP; black arrow) and signal peptide peptidase (SPP; gray arrow), to yield core, E1, E2, and p7. Two virus-encoded proteases, the NS2-NS3 autoprotease (orange arrowhead) and the NS3 serine protease as a complex with NS4A (purple arrowhead), generate mature proteins from the NS2–NS5B region. Core, E1, and E2 provide physical components of the virion. p7 and NS2 appear to function exclusively in and are essential for virus assembly (red stars). NS4A and NS5B (black circles) are critical for viral RNA replication, but there are no reports suggesting that either protein is required for generation of infectious progeny. NS3, NS4B, and NS5A have dual functions in viral RNA replication and particle production (blue diamonds).