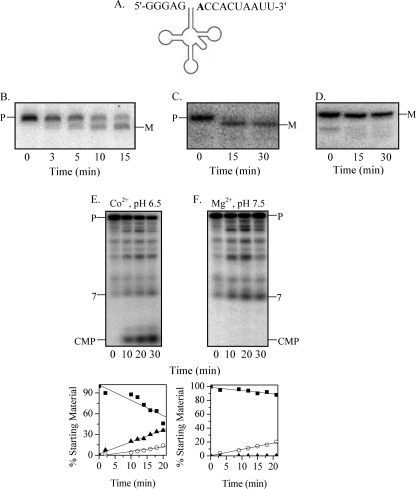
FIGURE 3.
Maturation of tRNAPheV precursor by RNase BN. A, shown is the structure of the E. coli tRNAPhe precursor. The discriminator nucleotide is shown in boldface. B, the tRNAPhe precursor (0.05 μm) with 6 extra 3′-residues after the CCA sequence was treated with RNase BN (0.14 μm), and the cleavage products were analyzed by 6% denaturing PAGE, followed by Northern blotting with a 5′-probe. C, the conditions were the same as described for B except that the precursor has 3 extra 3′-nt. D, the conditions were the same as described for B except that the substrate was mature (M) tRNAPhe. E and F, tRNAPhe labeled with [32P]pC at its 3′-end (48 nm) was treated with RNase BN (0.14 μm) in the presence of either Co2+ at pH 6.5 (E) or Mg2+ at pH 7.5 (F). The cleavage products were analyzed by 20% denaturing PAGE. Quantitation of the data from experiments similar to those in E and F is shown below the gels. Disappearance of the precursor (P) (■) and generation of CMP (▴) and 7-nt oligomer (○) products are presented as a percentage of the initial amount of precursor.