Abstract
Receptor for advanced glycation end product (RAGE)-dependent signaling has been implicated in ischemia/reperfusion injury in the heart, lung, liver, and brain. Because macrophages contribute to vascular perturbation and tissue injury in hypoxic settings, we tested the hypothesis that RAGE regulates early growth response-1 (Egr-1) expression in hypoxia-exposed macrophages. Molecular analysis, including silencing of RAGE, or blockade of RAGE with sRAGE (the extracellular ligand-binding domain of RAGE), anti-RAGE IgG, or anti-AGE IgG in THP-1 cells, and genetic deletion of RAGE in peritoneal macrophages, revealed that hypoxia-induced up-regulation of Egr-1 is mediated by RAGE signaling. In addition, the observation of increased cellular release of RAGE ligand AGEs in hypoxic THP-1 cells suggests that recruitment of RAGE in hypoxia is stimulated by rapid production of RAGE ligands in this setting. Finally, we show that mDia-1, previously shown to interact with the RAGE cytoplasmic domain, is essential for hypoxia-stimulated regulation of Egr-1, at least in part through protein kinase C βII, ERK1/2, and c-Jun NH2-terminal kinase signaling triggered by RAGE ligands. Our findings highlight a novel mechanism by which an extracellular signal initiated by RAGE ligand AGEs regulates Egr-1 in a manner requiring mDia-1.
Keywords: Hypoxia, Macrophage, Mouse Genetics, Signal Transduction, siRNA, Transcription Factors, AGE, Egr-1, RAGE
Introduction
The occlusion of blood vessels or insufficient blood flow to diseased tissues occurs with the onset and progression of many pathological states (1–6). Macrophages accumulate in large numbers in such ischemic/hypoxic areas and respond to oxygen signaling mechanisms involving a number of transcription factors (7–9). One such transcription factor, early growth response-1 (Egr-1),2 an inducible zinc finger transcription factor, is rapidly up-regulated in macrophages in coordinating inflammatory and procoagulant response to hypoxia (8, 9).
The generation of advanced glycation end products (AGEs) has been implicated in ischemia/reperfusion injury in the heart (10–14). AGE-modified proteins are able to activate macrophages and stimulate secretion of cytokines and inflammatory factors (15–17). A major mechanism by which AGEs exert their cellular effects is by ligation of the multiligand receptor for AGE (RAGE) (18). In addition, our previous findings demonstrated that AGE-RAGE-dependent membrane translocation of protein kinase C (PKC) βII and consequent activation of JNK signaling in the heart and in endothelial cells subjected to hypoxia directly impact on regulation of Egr-1 (19). However, ligands of RAGE are not simply tethered to this receptor.
Studies in vivo and in vitro revealed that cytoplamic domain of RAGE is essential for RAGE ligand-triggered signal transduction because deletion of the cytoplasmic domain of RAGE blocks ligands from inducing signaling and modulating gene expression (20). In addition, the cytoplasmic domain of RAGE interacts with a member of the formin homology domain proteins, diaphanous or mDia-1, which has been identified as a binding partner of the RAGE cytoplasmic domain (21). Previous experiments in RAGE-expressing transformed cells revealed that reduction of mDia-1 expression by small interfering RNAs (siRNA) significantly blocked the effects of RAGE ligands on cellular migration and activation of rac-1 and cdc42 (21).
In this study, we tested the hypothesis that RAGE regulates Egr-1 expression in hypoxia-exposed macrophages and that RAGE-dependent regulation of Egr-1 requires mDia-1 in these cells. Two types of macrophages were used as our experimental cell models: (i) the human monocytic leukemia cell line, THP-1, which shares many properties with human monocyte-derived macrophages (22, 23), is a useful model for studying the effects of RAGE signaling on regulation of Egr-1 expression by introduction of siRNA to knock down RAGE or mDia-1; (ii) murine peritoneal macrophages are useful models for studying the effects of RAGE signaling on regulation of Egr-1 by genetic ablation of RAGE or mDia-1. Our studies elucidate a novel signaling pathway by which RAGE-ligand stimulated up-regulation of Egr-1 in macrophages requires mDia-1.
EXPERIMENTAL PROCEDURES
Cell Culture and Induction of Hypoxia
A human monocytic leukemia cell line, THP-1, was obtained from the American Type Culture Collection (ATCC) and maintained in suspension in T-75 culture flasks at a cell density of 2.0 × 105/ml to 1.0 × 106/ml in 50 m1 of RPMI 1640 medium (ATCC) containing 10% fetal bovine serum, 10 mm HEPES, 1 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mm sodium pyruvate, and 50 μm β-mercaptoethanol. Cells were seeded in T-25 flasks at a density of 0.5 × 106 cells/ml or 0.8 × 106 cells/ml for preparation of RNA or nuclear extracts, respectively.
Mouse peritoneal macrophages were prepared from three groups of mice in our laboratory. C57BL/6 mice were purchased from Jackson Laboratories and used as wild-type control. RAGE−/− mice were bred in house. The DrfI−/− mice (DrfI gene targeted mDia-1 mice because the DrfI gene encodes the canonical formin mDia-1) were a gift from Dr. Alberts (24), and their offspring were produced in house. Both RAGE−/− and DrfI−/− mice were backcrossed >10 generations into C57BL/6 prior to study. These three groups of mice at age 8 weeks old or older were subjected to intraperitoneal inoculation of sterile 10% thioglycollate broth (Difco Laboratories) (25). On day 5 after injection, mice were anesthetized with isoflurane prior to rapid cervical dislocation. Approximately 10 ml of ice-cold serum-free Dulbecco's modified Eagle's medium was injected intraperitoneally through the exposed peritoneum. After a gentle massage, peritoneal fluid-containing cells were harvested and centrifuged at 3,000 rpm for 10 min. The cell pellet was suspended in 1 ml of red blood cell lysis buffer for 10–30 min to lyse contaminating erythrocytes and washed with phosphate-buffered saline twice and resuspended at a concentration of 1.5–2.0 × 106 cells/ml in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum plus 100 units/ml penicillin and 100 μg/ml streptomycin. The cell suspension was plated onto 35-mm tissue culture dishes at 2 ml/dish and incubated overnight at 37 °C in a humidified 5% CO2 atmosphere. Nonadherent cells were removed by replacing the complete growth medium, and the cultures were then incubated for 24 h at 37 °C in a humidified 5% CO2 atmosphere. The cells were replaced with serum-free medium for 24 h and exposed to normoxia or hypoxia.
Cells were subjected to hypoxia in an In Vivo 400 hypoxic work station (Biotrace, Cincinnati, OH), where cells were immediately immersed in the oxygen-deprived environment by replacing medium preequilibrated with the hypoxic gas mixture, and the incubation proceeded in atmosphere containing 5% CO2, 0.5% O2, and a balance of nitrogen. For in vitro inhibition analyses, cells were seeded in 35-mm dishes and preincubated with soluble (s)RAGE (25 μg/ml), anti-AGE IgG (15 μg/ml), anti-RAGE IgG (15 μg/ml), or aminoguanidine (50 and 200 μm) for 2 h (26), inhibitors of PKCβ (LY379196, 30 nm), ERK1/2 (U0126, 5 μm), JNK (SP600125, 20 μm), or p38 (SB203580, 20 μm) for 45 min and then subjected to hypoxia. Triplicate dishes from both the experimental hypoxic cultures and the control ambient cultures were processed at the indicated times after the start of hypoxic treatment. The experiments were carried out three times, in an identical fashion. Cells were harvested for the assays described below.
siRNA to Knock Down RAGE and mDia-1
siRNA duplexes against human RAGE (ID nos. 110857 and 110859) or against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Ambion. The mDia-1 siRNA was synthesized by Invitrogen. The siRNA duplexes against RAGE, mDia-1, or GAPDH were electroporated into THP-1 cells using the Nucleofector reagent and device according to the manufacturer's protocol (Lonza). To control for off-target effects of siRNA, a separate well of THP-1 cells was electroporated with a scramble siRNA as a negative control. After 6 h, cells were subjected to hypoxia followed by RNA isolation or protein preparation as described below.
RNA Extraction and Real-time Quantitative PCR
Total RNA was extracted from THP-1 cells or mouse peritoneal macrophages using the RNeasy Plus Mini kit (Qiagen). Total RNA (0.5 μg) was processed directly to cDNA synthesis using the TaqMan Reverse Transcription Reagents kit (Applied Biosystems) according to the manufacturer's protocol. Real-time PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems). All PCR primers and TaqMan probes were designed using software PrimerExpress (Applied Biosystems) and published sequence data from the NCBI database. The sequences of forward and backward primers and probe for human Egr-1 are 5′-GTCCCCGCTGCAGATCTCT-3′ and 5′-CTCCAGCTTAGGGTAGTTGTCCAT-3′, which were synthesized from Applied Biosystems. The sequence of TaqMan probe for human Egr-1 is 5′-CGGATCCTTTCCTCACTCGCCCA-3′, which was labeled with the reporter dye 6FAMTM in the 5′ end and minor groove binder with a nonfluorescent quencher in the 3′ end from Applied Biosystems. The sequences of primers and probe for mouse Egr-1 were same as previously published (27). The primers and probe for 18 S rRNA were purchased from Applied Biosystems. All reactions were performed in triplicate in an ABI PRISM 7900HT Sequence Detection system; 18 S rRNA was used as an active and endogenous reference to correct for differences in the amount of total RNA added to the reaction and to compensate for different levels of inhibition during reverse transcription of RNA and during PCR. Data are calculated by the 2−ΔΔCT method (28) and are presented as the fold induction of mRNA for Egr-1 in hypoxia-stimulated THP-1 to 18 S rRNA compared with nomoxic THP-1 cells (defined as 1.0-fold).
Western Blotting
Nuclear extracts and membrane protein were prepared from THP-1 cells as previously described (8, 9). Total protein extracts were prepared from THP-1 cells using cell lysis buffer (Cell Signaling Technology, Beverly, MA). Protein concentration was determined using the Bio-Rad protein assay. Equal amounts of protein (5, 30, 50 μg/sample) were subjected to SDS-PAGE (4–12%) followed by electrophoretic transfer to nitrocellulose membranes. Nonspecific binding was blocked by incubation of membranes with nonfat dry milk (5%) for 1 h at room temperature or overnight at 4 °C. The blot was incubated with one of the following antibodies as the primary antibody for the reaction: rabbit anti-Egr-1 IgG, anti-phospho-ERK1/2 (P-ERK1/2) IgG, anti-total-ERK1/2 (T-ERK1/2) IgG, anti-phospho-JNK (P-JNK) IgG, anti-total-JNK (T-JNK) IgG (Cell Signaling Technology), anti-Sp1 IgG, anti-PKCβII IgG (Santa Cruz Biotechnology Inc., Santa Cruz, CA), and anti-mDia-1 IgG (Abcam). Each primary antibody was used at a dilution of 1:1,000 for 1–3 h or overnight according to the manufacturer's instructions. Horseradish peroxidase-conjugated secondary antibodies (1:2,000; Amersham Biosciences or Sigma) were used to identify sites of binding of each primary antibody.
Electrophoretic Mobility Gel Shift Assay and Antibody Supershift Assay
Nuclear extracts were prepared from THP-1 cells, and electrophoretic mobility gel shift assay was performed as described (8, 9). For antibody supershift assays, nuclear extracts were preincubated with anti-Egr-1 IgG (Santa Cruz Biotechnology) and then subjected to binding reaction (8, 9).
ELISA for Detection of AGE and High Mobility Group Box 1 (HMGB1)
After THP-1 cells were serum-starved for 24 h and then subjected to hypoxia as described above, supernatants were harvested and stored at −80 °C. Cell lysates were prepared using cell lysis buffer (Cell Signaling Technology) for determining protein concentrations. To test AGE, equal amounts of protein were coated overnight onto an ELISA plate using carbonate-bicarbonate buffer (Sigma). AGE ELISA was performed using T-gel (Pierce) affinity-purified chicken anti-AGE as the primary antibody (10, 19) at a concentration of 30 μg/ml for 3 h at room temperature. The secondary antibody (anti-chicken IgY) (Sigma) was diluted to 1:10,000 for 1 h at room temperature. The signals were developed in phosphate-citrate (Sigma) and hydrogen peroxide (Sigma). Ribose-glycated albumin was used to prepare the standard curve (10, 19). To test HMGB1, the assay was performed using a HMGB1 ELISA kit according to the manufacturer's instructions (IBL International GMBH).
Statistical Analysis
All data are expressed as the mean ± S.E. Statistical significance was analyzed by analysis of variance using commercially available (Statview, version 5.0.1, Berkeley, CA) software. p values < 0.05 were considered statistically significant.
RESULTS
Hypoxia-induced Egr-1 Expression in THP-1 Cells
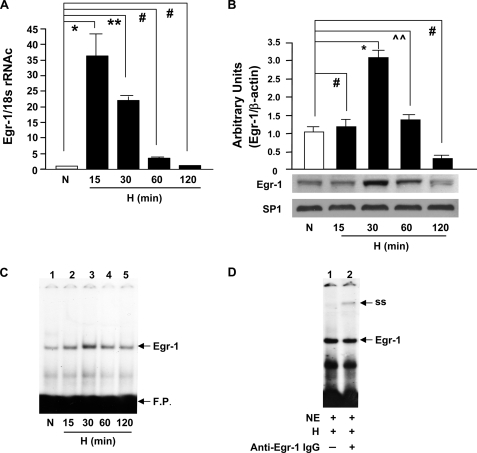
Human THP-1 cells were subjected to hypoxia (O2, 0.5%) using an In Vivo 400 hypoxic work station. Real-time PCR analysis revealed that in THP-1 cells, Egr-1 transcripts increased in a time-dependent manner, with a maximum expression at 15 min of hypoxia versus normoxia (≈31-fold, *, p < 0.0001; Fig. 1A). By 60 and 120 min of hypoxia, no significant increase in Egr-1 mRNA transcripts versus normoxia was noted in THP-1 cells (Fig. 1A). Immunoblotting of nuclear extracts prepared from THP-1 cells displayed an ≈4-fold increase in Egr-1 nuclear protein at 30 min of hypoxia compared with normoxia (Fig. 1B; *, p < 0.0001). In contrast to the observed induction of Egr-1, there was no increase in Sp1 in hypoxic THP-1 cells. Electrophoretic mobility shift analysis of nuclear extracts from THP-1 cells using 32P-labeled consensus oligonucleotide probe for Egr DNA binding activity demonstrated a gel shift band whose intensity increased in a time-dependent manner, with a maximum shift at 30 min of hypoxia compared with normoxia (Fig. 1C). That this band in hypoxic THP-1 cells contained Egr-1 protein was illustrated by the presence of a supershifted band shown in nuclear extracts treated with an anti-Egr-1 antibody (denoted as ss in Fig. 1D).
FIGURE 1.
Hypoxia-mediated induction of Egr-1 in THP-1 cells. THP-1 cells were exposed to hypoxia (H; 0.5% of oxygen) or normoxia (N) for the indicated times, and total RNA or nuclear extracts were isolated. A, real-time PCR analysis of Egr-1 expression was performed. Data are represented as the relative expression of mRNA for Egr-1 normalized to 18 S rRNA. B, immunoblotting with anti-Egr-1 IgG was performed on 5 μg/lane of nuclear protein from THP-1 cells, and results of multiple experiments were quantified. C, electrophoretic mobility gel shift assay was performed with 32P-labeled consensus Egr-1 oligonucleotide probe and nuclear extracts (3 μg/lane of nuclear protein) from THP-1 cells. D, supershift assay using anti-Egr-1 antibody was performed with 32P-labeled consensus Egr-1 oligonucleotide probe and nuclear extracts (3 μg/lane of nuclear protein) from THP-1 cells after 30 min of hypoxia. All experiments were repeated more than three times; representative bands are shown, and the mean ± S.E. (error bars) is shown. * (p < 0.0001), ** (p < 0.001), and ⋀⋀ (p < 0.05) indicate statistical significance; # indicates no statistical significance. F. P. indicates free probe. NE indicates nuclear extract.
Essential Role for RAGE in Hypoxia-mediated Expression of Egr-1 in Macrophages
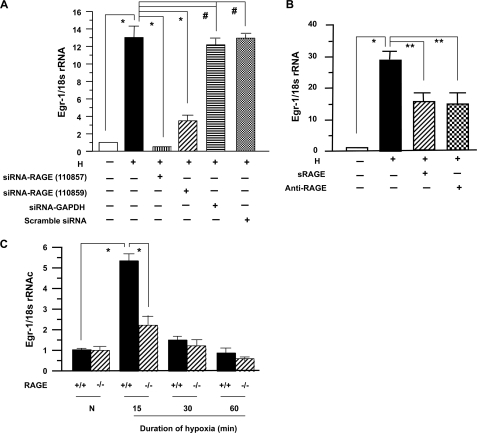
To probe the potential role of RAGE in impacting up-regulation of Egr-1 in hypoxic macrophages, we employed a number of distinct strategies. First, we introduced siRNA to reduce RAGE expression in THP-1 cells exposed to hypoxia. Introduction of siRNA to knock down RAGE expression by either 110857 or 110859 markedly blunted the effect of hypoxia on up-regulation of Egr-1 transcripts (Fig. 2A; *, p < 0.0001), although the efficiency of blockade by 110857 was somewhat higher than that of 110859. Neither of them had any effect on expression of 18 S rRNA (Fig. 2A). Introduction of siRNA against GAPDH or scrambled siRNA had no effect on up-regulation of Egr-1 in THP-1 cells in hypoxia (Fig. 2A). Consistent with these findings, treatment of THP-1 cells with anti-RAGE IgG or sRAGE, the extracellular ligand-binding domain of RAGE, reduced hypoxia-mediated up-regulation of Egr-1 transcripts (Fig. 2B; **, p < 0.001).
FIGURE 2.
RNA interference silencing, pharmacological blockade, or genetic deletion of RAGE attenuates hypoxia-mediated expression of Egr-1 in THP-1 cells and macrophages. THP-1 cells and peritoneal Mϕ from RAGE+/+ and RAGE−/− mice were exposed to hypoxia (H; 0.5% of oxygen) or normoxia (N) for the indicated times. Total RNA was prepared from those cells, and real-time PCR analysis of Egr-1 expression was performed. Data are represented as the relative expression of mRNA for Egr-1 normalized to 18 S rRNA. A, RNA was prepared from THP-1 cells transfected with or without siRNA-RAGE, scramble siRNA, or siRNA-GADPH and subjected to hypoxia for 15 min. B, RNA was prepared from THP-1 cells preincubated with or without sRAGE (25 μg/ml) or anti-RAGE IgG (15 μg/ml) and then subjected to hypoxia for 15 min. C, RNA was prepared from RAGE+/+ and RAGE−/− Mϕ after exposure to hypoxia or normoxia for the indicated times. * (p < 0.0001) and ** (p < 0.001) indicate statistical significance; #, no statistical significance.
Next, we determined whether genetic deletion of RAGE affected up-regulation of Egr-1 in hypoxic macrophages. Elicited mouse peritoneal macrophages (Mϕ) by thioglycollate were prepared from RAGE+/+ and RAGE−/− mice. After they were cultured for 2 days and serum-starved for 24 h, they were subjected to hypoxia (O2, 0.5%). Real-time PCR analysis revealed that in RAGE+/+ Mϕ, Egr-1 transcripts were increased ≈5.3-fold at 15 min of hypoxia versus normoxia (Fig. 2C; *, p < 0.0001). In contrast, RAGE−/− Mϕ subjected to hypoxia for the same time showed only an ≈2.3-fold increase in Egr-1 transcripts compared with normoxia (Fig. 2C). Expression level of Egr-1 transcripts in RAGE−/− Mϕ was ≈2.4-fold lower than that in RAGE+/+ Mϕ at 15 min of hypoxia (Fig. 2C; *, p < 0.0001). However, the level of expression of Egr-1 transcripts in both RAGE+/+ and RAGE−/− Mϕ declined during 30–60 min of exposure to hypoxia versus normoxia (Fig. 2C).
Hypoxia-induced mDia-1 Expression and Its Impact on Up-regulation of Egr-1 in THP-1 Cells and Murine Macrophages
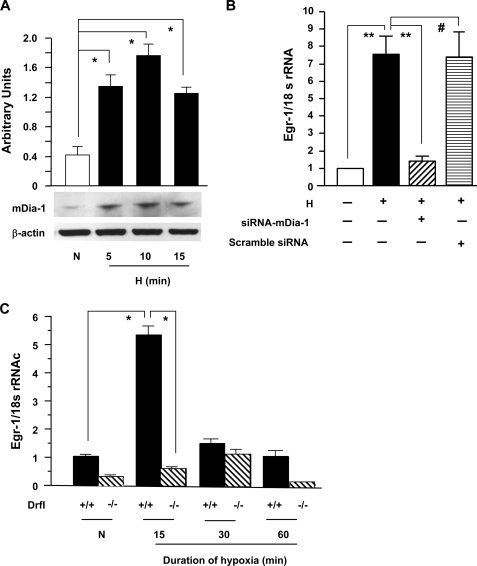
Because of the previous findings that the RAGE cytoplasmic domain interacts with diaphanous-1 (mDia-1), a member of the formin homology domain protein family (21), and that mDia-1 was required for RAGE ligand-stimulated cellular migration and activation of rac-1 and cdc42 in transformed cells (21), we sought to test whether mDia-1 was expressed in THP-1 cells and whether expression was enhanced in THP-1 cells exposed to hypoxia. As shown in Fig. 3A, immunoblotting of total protein lysates revealed significantly higher expression of mDia-1 in THP-1 cells exposed to hypoxia for 5, 10, or 15 min versus normoxia (≈4.5-fold; *, p < 0.0001; Fig. 3A). Second, to test whether mDia-1 was important in RAGE-dependent-up-regulation of Egr-1 expression in hypoxia, siRNA against mDia-1 or scramble siRNA was applied to THP-1 cells. Introduction of siRNA to knock down mDia-1 expression significantly blunted the effect of hypoxia on up-regulation of Egr-1 transcripts (Fig. 3B; **, p < 0.001) but had no effect on expression of 18 S rRNA (Fig. 3B). Introduction of scrambled siRNA had no effect on up-regulation of Egr-1 in THP-1 cells exposed to hypoxia (Fig. 3B). Next, we determined whether genetic deletion of mDia-1 affected up-regulation of Egr-1 in hypoxic macrophages. Mouse Mϕ elicited by thioglycollate were prepared from DrfI+/+ and DrfI−/− mice. After they were cultured for 2 days and serum-starved for 24 h, they were subjected to hypoxia (O2, 0.5%). Levels of Egr-1 transcripts in wild-type DrfI+/+ Mϕ were much higher (≈9.2-fold) than those in DrfI−/− Mϕ at 15 min of hypoxia (Fig. 3C; *, p < 0.0001). These data confirmed that mDia-1 was required for hypoxia-stimulated up-regulation of Egr-1 in Mϕ.
FIGURE 3.
Hypoxia-mediated induction of mDia-1 in THP-1 cells and its impact on up-regulation of Egr-1 in THP-1 cells and macrophages. THP-1 cells and Mϕ from mDia-1 DrfI+/+ and DrfI−/− mice were exposed to hypoxia (H; 0.5% of oxygen) or normoxia (N) for the indicated times. A, total protein was prepared from THP-1 cells, and immunoblotting with anti-mDia-1 IgG was performed on 30 μg/lane of protein from THP-1 cells. Results of multiple experiments were quantified. B and C, total RNA was prepared from those cells, and real-time PCR analysis of Egr-1 expression was performed. Data are represented as the relative expression of mRNA for Egr-1 normalized to 18 S rRNA. B, RNA was prepared from THP-1 cells transfected with siRNA-mDia-1 or scramble siRNA and then subjected to hypoxia for 15 min. C, RNA was prepared from DrfI+/+ and DrfI−/− Mϕ after exposure to hypoxia or normoxia for the indicated times. * (p < 0.0001) and ** (p < 0.001) indicate statistical significance; #, no statistical significance.
Hypoxia-induced Signal Transduction Pathways Mediating Egr-1 Regulation in THP-1 Cells
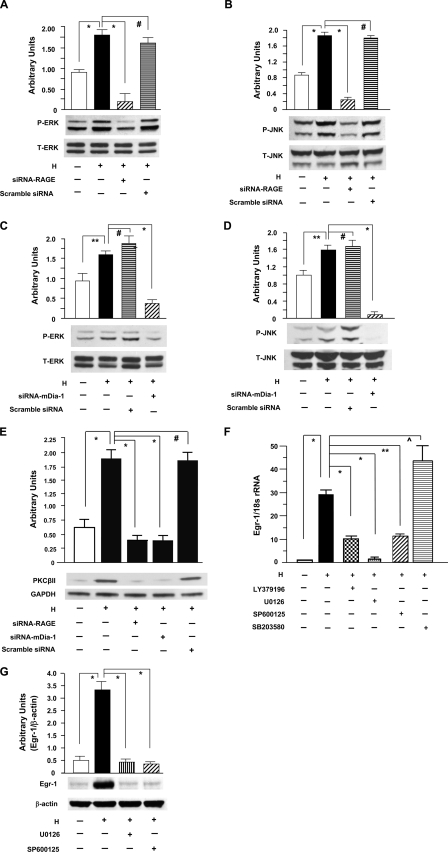
In view of roles for RAGE and downstream mitogen-activated protein kinases in environmental stress and regulation of Egr-1 (11, 18, 19, 29–32), we investigated the specific impact of RAGE on ERK1/2 and JNK activation in hypoxia. Cultured THP-1 cells were exposed to hypoxia for the indicated time points. Immunoblotting of extracts from hypoxic THP-1 cells displayed an increase in intensity of phospho-ERK1/2 and phospho-JNK in a time-dependent manner (data not shown), with peak activation at 10-min exposure to hypoxia, compared with extracts from normoxic THP-1 cells (Fig. 4, A and C, and Fig. 4, B and D; *, p < 0.0001). Next, we tested whether RAGE and mDia-1 are required for activation of ERK1/2 or JNK in hypoxic THP-1 cells. Introduction of siRNA to knock down RAGE expression in THP-1 cells blunted the effect of hypoxia on phosphorylation of ERK1/2 (Fig. 4A; *, p < 0.0001) and JNK (Fig. 4B; *, p < 0.0001). In parallel, silencing of mDia-1 by introducing siRNA against mDia-1 in THP-1 cells blocked phosphorylation of ERK1/2 (Fig. 4C; *, p < 0.0001) and JNK (Fig. 4D; *, p < 0.0001) in response to hypoxia. However, introduction of scrambled siRNA had no effect of hypoxia on activation of ERK1/2 and JNK in THP-1 cells (Fig. 4, A and C, and Fig. 4, B and D). Levels of total ERK1/2 and JNK proteins did not change in each of these experiments (Fig. 4, A–D). These data confirmed that both RAGE and mDia-1 were essential for activation of ERK1/2 and JNK in hypoxic THP-1 cells.
FIGURE 4.
Hypoxia-mediated activation of PKCβII, ERK1/2, and JNK leading to up-regulation of Egr-1 in THP-1 cells. Total protein, membrane protein, nuclear protein, and RNA were prepared from THP-1 cells exposed to hypoxia (H; 0.5% of oxygen) or normoxia (N) for the indicated times. A–D, immunoblotting with anti-phospho-ERK1/2 (P-ERK1/2) IgG (A and C) and anti-total-ERK1/2 (T-ERK1/2) IgG (A and C) or with anti-phospho-JNK (P-JNK) IgG (B and D) and anti-total-JNK (T-JNK) (B and D) was performed on 5 or 50 μg/lane of protein from THP-1 cells transfected with or without siRNA-RAGE (A and B) or siRNA-mDia-1 (C and D) and scramble siRNA and subjected to hypoxia for 10 min. E, immunoblotting with anti-PKCβII IgG was performed on 5 μg/lane of membrane protein from THP-1 cells transfected with or without siRNA-RAGE or siRNA-mDia-1 and scramble siRNA and subjected to hypoxia for 10 min. F and G, RNA and nuclear protein were prepared from THP-1 cells preincubated with or without inhibitors of PKCβ (LY379196, 30 nm), ERK1/2 (U0126, 5 μm), JNK (SP600125, 20 μm), and p38 (SB203580, 20 μm) and subjected to hypoxia for 15 (F) or 30 (G) min. F, real-time PCR analysis of Egr-1 expression was performed. Data are represented as the relative expression of mRNA for Egr-1 normalized to 18 S rRNA. G, immunoblotting with anti-Egr-1 IgG was performed on 5 μg/lane of nuclear protein from those THP-1 cells. All results of multiple experiments were quantified. * (p < 0.0001), ** (p < 0.001), and ⋀ (p < 0.05) indicate statistical significance; #, no statistical significance.
Because our previous findings demonstrated significantly increased PKCβII antigen in the membranous fraction and PKCβ-dependent regulation of phosphorylation of ERK1/2 in hypoxic macrophages (9) and PKCβ-dependent regulation of phosphorylation of JNK in hypoxic endothelial cells (19), we sought to detect the impact of RAGE or mDia-1 on activation of PKCβII in THP-1 cells after hypoxia. An increased protein level of PKCβII in membranous fractions in THP-1 cells was observed at the maximum 10-min exposure to hypoxia compared with normoxic controls (Fig. 4E; *, p < 0.0001). In contrast, silencing of RAGE or mDia-1 by introduction of siRNA in THP-1 cells diminished hypoxia-induced activation of PKCβII in THP-1 cells after hypoxia (Fig. 4E; *, p < 0.0001). Loading controls using anti-GAPDH IgG indicated identical protein loading (Fig. 4E).
In view of the striking effects of anti-RAGE IgG and sRAGE on suppression of RAGE-dependent Egr-1 up-regulation in THP-1 cells exposed to hypoxia, we next directly tested the effects of the inhibitors of PKCβ and mitogen-activated protein kinase on Egr-1 expression. Real-time PCR analysis of total RNA from THP-1 cells demonstrated that the robust increase in Egr-1 transcripts in hypoxia was strongly suppressed by inhibitors of PKCβ (LY3979196, 30 nm), ERK1/2 (U0126, 5 μm), and JNK (SP600125, 20 μm) (Fig. 4F; *, p < 0.0001); but an inhibitor of p38 (SB203580, 10 μm) had no suppressive effect on hypoxia-stimulated induction of Egr-1 transcripts (Fig. 4F). In parallel, immunoblotting analysis of nuclear protein from THP-1 cells revealed that the robust increase in Egr-1 nuclear protein in hypoxia was strongly suppressed by inhibitors of ERK1/2 and JNK (Fig. 4G; *, p < 0.0001). These findings suggested that hypoxia-mediated Egr-1 up-regulation was accounted for by activation of PKCβII, phosphorylation of ERK1/2 and JNK, but not via phosphorylation of p38 in THP-1 cells in response to hypoxia.
Mechanisms by Which RAGE Regulates Egr-1 Expression in Hypoxic THP-1 Cells
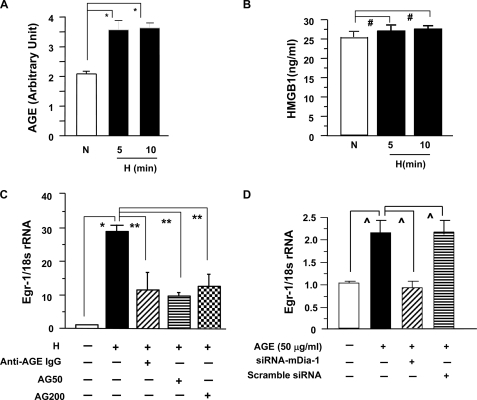
Our previous findings demonstrated that hypoxia generated one of the classes of RAGE ligands, AGEs, in endothelial cells (19). We measured these species in THP-1 supernatants in normoxia or after oxygen deprivation. Increased AGE-immunoreactive epitopes were detected in supernatants of THP-1 cells by ELISA and achieved significance at 5 and 10 min of hypoxia versus normoxia control (Fig. 5A; *, p < 0.0001). Because RAGE is a multiligand receptor, we investigated whether hypoxia induced release of non-AGE ligands, such as HMGB1. No significant changes of HMGB1 by ELISA were observed in supernatants of THP-1 cells at 5- and 10-min exposure to hypoxia compared with normoxia (Fig. 5B).
FIGURE 5.
AGE-mediated up-regulation of Egr-1 in THP-1 cells: dependence on RAGE and mDia-1. A and B, THP-1 cells were subjected to hypoxia for the indicated times. Supernatants were harvested and applied to ELISA for detection of AGE (A) and HMGB1 (B) epitopes. C, THP-1 cells were preincubated with or without anti-AGE IgG (15 μg/ml), or aminoguanidine (AG) (50 and 200 μm) for 2 h and then subjected to hypoxia for 15 min. RNA was isolated from those THP-1 cells for analysis of Egr-1 expression by real-time PCR. D, THP-1 cells were transfected with siRNA-mDia-1 or scramble siRNA and then exposed to CML-AGE (50 μg/ml) for 15 min. RNA was isolated from those THP-1 cells for analysis of Egr-1 expression by real-time PCR. * (p < 0.0001), ** (p < 0.001), and ⋀ (p < 0.01) indicate statistical significance; #, no statistical significance.
Next, to implicate AGE-RAGE directly in the downstream events leading to up-regulation of Egr-1 in hypoxia, we used specific reagents, aminoguanidine or anti-AGE IgG according to our previous studies (19), to block AGE and its interaction with RAGE in THP-1 cells exposed to hypoxia. Treatment of THP-1 cells with aminoguanidine or anti-AGE IgG, diminished hypoxia-mediated up-regulation of Egr-1 (Fig. 5C; **, p < 0.001).
Finally, we directly probed the role of mDia-1 in RAGE ligand-dependent up-regulation of Egr-1. THP-1 cells, in the absence or presence of siRNA against mDia-1 or scramble siRNA, were exposed to the specific RAGE ligand CML-HSA (Nϵ-(carboxymethyl)lysine-human serum albumin) (50 μg/ml), a major antigenic AGE recognized by RAGE, for 15 min. Silencing of mDia-1, but not scramble control, markedly blocked CML AGE-stimulated up-regulation of Egr-1 transcripts (Fig. 5D; ⋀, p < 0.01). These data demonstrated that mDia-1 was required for RAGE ligand CML-AGE-stimulated up-regulation of Egr-1.
DISCUSSION
Because endothelial cells and macrophages contribute to vascular perturbation in tissue dysfunction and failure in hypoxia/ischemia, our investigation into mechanisms by which hypoxia may trigger macrophage-RAGE signaling in Egr-1 regulation was prompted by our previous findings that regulation of Egr-1 in the heart and in endothelial cells subjected to hypoxia is mediated by RAGE (19). In the present studies, first, we observed that hypoxia stimulates rapid production and cellular release of AGEs by THP-1 cells exposed to hypoxia. We confirmed that non-AGE ligand HMGB1 was not significantly elevated in THP-1 supernatants over the same time course in which RAGE-dependent mechanisms regulated Egr-1 in hypoxia. These data suggest that hypoxia-stimulated AGE production was the chief mechanism by which RAGE was engaged in hypoxic macrophages. Perhaps longer periods of hypoxia are necessary to stimulate release of HMGB1 because other studies demonstrated that hypoxia-induced HMGB1 release in the supernatants of hepatocytes was initiated at 8 h and reached the maximum at 24 h (33). Second, we provided evidence that regulation of Egr-1 in macrophages in response to hypoxia is mediated by AGE-RAGE signaling, based on the effects of down-regulation of RAGE expression by RNA interference, blockade of AGE-RAGE in THP-1 cells, or genetic deletion in thioglycollate-elicited Mϕ on AGE-RAGE-mediated up-regulation of Egr-1 in response to hypoxia. Beyond this, here we show for the first time that hypoxia- and AGE-induced macrophage-RAGE signaling in Egr-1 regulation requires the RAGE cytoplasmic domain binding partner, mDia-1, because down-regulation of mDia-1 expression by RNA interference in THP-1 cells or genetic deletion in thioglycollate-elicited peritoneal macrophages blocks AGE-RAGE-mediated up-regulation of Egr-1 in response to hypoxia. Importantly, silencing of mDia-1 markedly blocked RAGE-ligand stimulated up-regulation of Egr-1 in THP-1 cells directly exposed to CML-AGEs.
The diverse nature of RAGE signaling and the finding that the RAGE cytoplasmic domain lacked endogenous tyrosine kinase activity strongly suggested that RAGE interacted with cytoplasmic binding partner(s) to trigger the recruitment of downstream effector pathways (21). Although RAGE ligands stimulate activation of a diverse array of signaling cascades, including PKC, ERK1/2, p38, and JNK, as has been shown by us and others (11, 18, 19, 29–32), there is no evidence to date to show that RAGE ligand-stimulated activation of PKCβII, ERK1/2, and JNK requires mDia-1. In the present studies we demonstrated that siRNA-mediated knockdown of RAGE or mDia-1 in THP-1 cells resulted in marked reduction of hypoxia-stimulated activation of PKCβII, ERK1/2, and JNK compared with controls. Further, our data directly implicate PKCβ, ERK1/2, and JNK in hypoxia-mediated up-regulation of Egr-1, as demonstrated by the suppressive effects of LY39196, U0126, and SP600125, respectively.
Hypoxia is a critical stimulus of accumulation of macrophages in diseased tissues, and previous studies illustrated that in these cells, Egr-1 is essential for up-regulation of plasminogen activator inhibitor-1 (34, 35) and tissue factor (8, 34, 36). Plasminogen activator inhibitor-1 and tissue factor link Egr-1 and hypoxia to increased deposition and fibrin and, thereby, amplification of tissue injury mechanisms. Although RAGE has been shown to mediate monocyte/macrophage chemotaxis and up-regulation of inflammatory factors, these studies implicate RAGE signaling for the first time in macrophage responses in hypoxia and suggest that recruitment of RAGE in hypoxia is stimulated by rapid production of AGEs in this setting.
In conclusion, our data for the first time reveal that mDia-1 is essential for hypoxia-stimulated activation of PKCβII, ERK1/2, and JNK signaling triggered by RAGE ligands and, consequently, regulation of Egr-1 in hypoxic macrophages. In this work, our findings highlight a novel mechanism by which an extracellular signal initiated by RAGE ligand AGEs regulates Egr-1 in a manner dependent on mDia-1.
Acknowledgments
We thank Dr. Arthur S. Alberts for providing DrfI−/− mice and Latoya Woods for excellent assistance in the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HL60901 through the NHLBI. This work was also supported by the U.S. Public Health Service and the Juvenile Diabetes Research Foundation.
- Egr
- early growth response
- AGE
- advanced glycation end product
- RAGE
- receptor for AGE
- CML
- Nϵ-(carboxymethyl) lysine
- ELISA
- enzyme-linked immunosorbent assay
- ERK
- extracellular signal-regulated kinase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HMBG
- high mobility group box
- JNK
- c-Jun NH2-terminal kinase
- Mϕ
- macrophages
- PKC
- protein kinase C
- RAGE
- receptor for AGE
- siRNA
- small interfering RNA
- sRAGE
- soluble RAGE.
REFERENCES
- 1.Blouw B., Song H., Tihan T., Bosze J., Ferrara N., Gerber H. P., Johnson R. S., Bergers G. (2003) Cancer Cell 4, 133–146 [DOI] [PubMed] [Google Scholar]
- 2.Björnheden T., Levin M., Evaldsson M., Wiklund O. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 870–876 [DOI] [PubMed] [Google Scholar]
- 3.Lambert J. M., Lopez E. F., Lindsey M. L. (2008) Int. J. Cardiol. 130, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens C. R., Williams R. B., Farrell A. J., Blake D. R. (1991) Ann. Rheum. Dis. 50, 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albina J. E., Reichner J. S. (2003) Wound Repair Regen. 11, 445–451 [DOI] [PubMed] [Google Scholar]
- 6.Ikeda E. (2005) Pathol. Int. 55, 603–610 [DOI] [PubMed] [Google Scholar]
- 7.Lewis J. S., Lee J. A., Underwood J. C., Harris A. L., Lewis C. E. (1999) J. Leukocyte Biol. 66, 889–900 [DOI] [PubMed] [Google Scholar]
- 8.Yan S. F., Zou Y. S., Gao Y., Zhai C., Mackman N., Lee S. L., Milbrandt J., Pinsky D., Kisiel W., Stern D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8298–8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan S. F., Lu J., Zou Y. S., Soh-Won J., Cohen D. M., Buttrick P. M., Cooper D. R., Steinberg S. F., Mackman N., Pinsky D. J., Stern D. M. (1999) J. Biol. Chem. 274, 15030–15040 [DOI] [PubMed] [Google Scholar]
- 10.Bucciarelli L. G., Kaneko M., Ananthakrishnan R., Harja E., Lee L. K., Hwang Y. C., Lerner S., Bakr S., Li Q., Lu Y., Song F., Qu W., Gomez T., Zou Y. S., Yan S. F., Schmidt A. M., Ramasamy R. (2006) Circulation 113, 1226–1234 [DOI] [PubMed] [Google Scholar]
- 11.Aleshin A., Ananthakrishnan R., Li Q., Rosario R., Lu Y., Qu W., Song F., Bakr S., Szabolcs M., D'Agati V., Liu R., Homma S., Schmidt A. M., Yan S. F., Ramasamy R. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1823–H1832 [DOI] [PubMed] [Google Scholar]
- 12.Bucciarelli L. G., Ananthakrishnan R., Hwang Y. C., Kaneko M., Song F., Sell D. R., Strauch C., Monnier V. M., Yan S. F., Schmidt A. M., Ramasamy R. (2008) Diabetes 57, 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan S. F., Ramasamy R., Schmidt A. M. (2009) Expert Rev. Mol. Med. 11, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brownlee M. (1992) Diabetes Care 15, 1835–1843 [DOI] [PubMed] [Google Scholar]
- 15.McCarthy A. D., Etcheverry S. B., Bruzzone L., Cortizo A. M. (1997) Mol. Cell. Biochem. 170, 43–51 [DOI] [PubMed] [Google Scholar]
- 16.Miyata T., Kawai R., Taketomi S., Sprague S. M. (1996) Nephrol. Dial. Transplant. 11, Suppl. 5, 54–57 [DOI] [PubMed] [Google Scholar]
- 17.Takagi M., Kasayama S., Yamamoto T., Motomura T., Hashimoto K., Yamamoto H., Sato B., Okada S., Kishimoto T. (1997) J. Bone Miner. Res. 12, 439–446 [DOI] [PubMed] [Google Scholar]
- 18.Kislinger T., Fu C., Huber B., Qu W., Taguchi A., Du Yan S., Hofmann M., Yan S. F., Pischetsrieder M., Stern D. M., Schmidt A. M. (1999) J. Biol. Chem. 274, 31740–31749 [DOI] [PubMed] [Google Scholar]
- 19.Chang J. S., Wendt T., Qu W., Kong L., Zou Y. S., Schmidt A. M., Yan S. F. (2008) Circ. Res. 102, 905–913 [DOI] [PubMed] [Google Scholar]
- 20.Yan S. F., Ramasamy R., Schmidt A. M. (2010) Biochem. Pharmacol. 79, 1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson B. I., Kalea A. Z., Del Mar Arriero M., Harja E., Boulanger E., D'Agati V., Schmidt A. M. (2008) J. Biol. Chem. 283, 34457–34468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T., Tada K. (1980) Int. J. Cancer 26, 171–176 [DOI] [PubMed] [Google Scholar]
- 23.Banka C. L., Black A. S., Dyer C. A., Curtiss L. K. (1991) J. Lipid Res. 32, 35–43 [PubMed] [Google Scholar]
- 24.Peng J., Kitchen S. M., West R. A., Sigler R., Eisenmann K. M., Alberts A. S. (2007) Cancer Res. 67, 7565–7571 [DOI] [PubMed] [Google Scholar]
- 25.Bianco C., Griffin F. M., Jr., Silverstein S. C. (1975) J. Exp. Med. 141, 1278–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies M. G., Hagen P. O. (1996) J. Surg. Res. 63, 474–479 [DOI] [PubMed] [Google Scholar]
- 27.Harja E., Bucciarelli L. G., Lu Y., Stern D. M., Zou Y. S., Schmidt A. M., Yan S. F. (2004) Circ. Res. 94, 333–339 [DOI] [PubMed] [Google Scholar]
- 28.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 29.Taguchi A., Blood D. C., del Toro G., Canet A., Lee D. C., Qu W., Tanji N., Lu Y., Lalla E., Fu C., Hofmann M. A., Kislinger T., Ingram M., Lu A., Tanaka H., Hori O., Ogawa S., Stern D. M., Schmidt A. M. (2000) Nature 405, 354–360 [DOI] [PubMed] [Google Scholar]
- 30.Yeh C. H., Sturgis L., Haidacher J., Zhang X. N., Sherwood S. J., Bjercke R. J., Juhasz O., Crow M. T., Tilton R. G., Denner L. (2001) Diabetes 50, 1495–1504 [DOI] [PubMed] [Google Scholar]
- 31.Harja E., Bu D. X., Hudson B. I., Chang J. S., Shen X., Hallam K., Kalea A. Z., Lu Y., Rosario R. H., Oruganti S., Nikolla Z., Belov D., Lalla E., Ramasamy R., Yan S. F., Schmidt A. M. (2008) J. Clin. Invest. 118, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J., Ananthakrishnan R., Qu W., Lu Y., Reiniger N., Zeng S., Ma W., Rosario R., Yan S. F., Ramasamy R., D'Agati V., Schmidt A. M. (2008) J. Am. Soc. Nephrol. 19, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsung A., Klune J. R., Zhang X., Jeyabalan G., Cao Z., Peng X., Stolz D. B., Geller D. A., Rosengart M. R., Billiar T. R. (2007) J. Exp. Med. 204, 2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan S. F., Fujita T., Lu J., Okada K., Shan Zou Y., Mackman N., Pinsky D. J., Stern D. M. (2000) Nat. Med. 6, 1355–1361 [DOI] [PubMed] [Google Scholar]
- 35.Liao H., Hyman M. C., Lawrence D. A., Pinsky D. J. (2007) FASEB J. 21, 935–949 [DOI] [PubMed] [Google Scholar]
- 36.Yan S. F., Mackman N., Kisiel W., Stern D. M., Pinsky D. J. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 2029–2035 [DOI] [PubMed] [Google Scholar]