Figure 1.
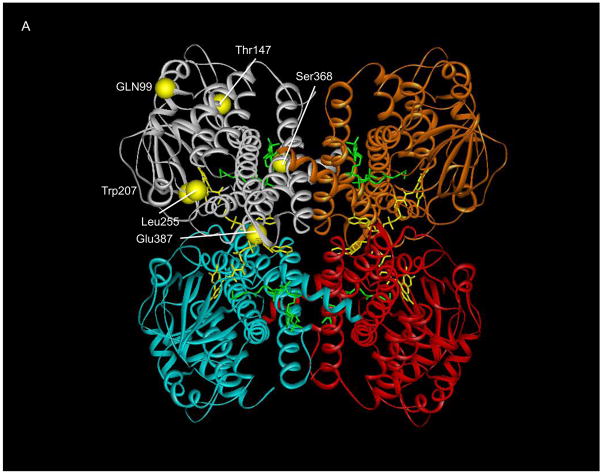
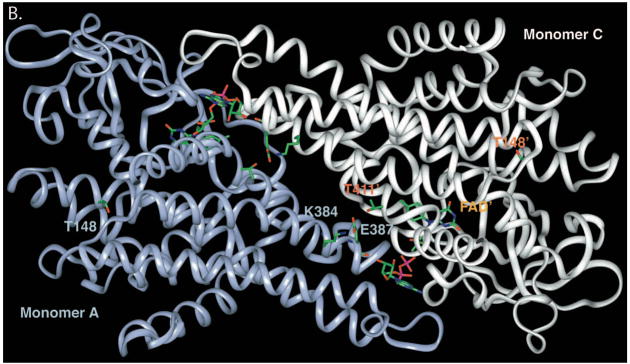
Ribbon representation of point mutations identified in SBCAD gene. A. The tetrameric SBCAD crystal structure, PDB code 2JIF, is shown with each subunit colored a different color. FAD is shown as yellow and bound substrate as green. The amino acid substitutions identified in this and other studies are represented as yellow balls with precursor protein numbering given. Leu255Phe and Ser368Pro have previously been reported in Korman, et al, 2005). The former is likely to be important in stabilizing the interaction between two adjacent alpha helices. Ser368 likes in an alpha helix that is likely to have its trajectory altered by substitution of a proline. B. Residues at the 148 and 387 positions (precursor numbering) are illustrated. The Thr148 hydroxlate group lies in a hydrophilic pocket, but does not directly interact with other residues side chains. The carboxylate oxygen molecules of Glu387 lie between the Lys384 side chain amino group and the hydroxyl group of Thr411’ of the second subunit forming a hydrogen bonding conduit. In addition, the Glu387 backbone oxygen is directly involved in FAD binding through hydrogen bonding.