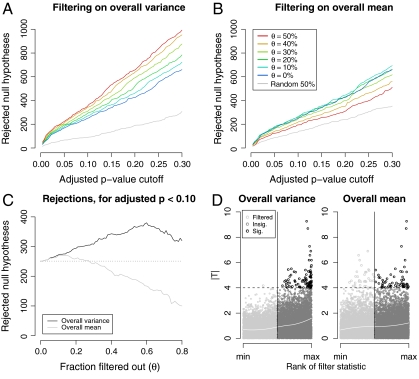
Fig. 1.
Power assessment of filtering applied to the ALL data (12,625 genes). R, the number of genes called differentially expressed between the two cytogenetic groups, was computed for different stage-one filters, filtering stringencies, and FDR-adjusted p-value cutoffs. In all cases, a standard t-statistic (T) was used in stage two, and adjustment for multiple testing was by the method of ref. 24. Similar results were obtained with other adjustment procedures. Filter cutoffs were selected so that a fraction θ of genes were removed. A random filter, which arbitrarily selected and removed one half of the genes, was also considered. (A) Filtering on overall variance (S2). At all FDR cutoffs, increasingly stringent filtering increased total discoveries, even though fewer genes were tested. This effect was not, however, due to the reduction in the number of hypotheses alone: filtering half of the genes at random reduced total discoveries by approximately one half, as expected. (B) Filtering on overall mean (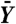 ), on the other hand, produced a small increase in rejections at low stringency, but then substantially reduced rejections, and thus power, at higher stringencies. (C) Effect of increasing filtering stringency for fixed adjusted p-value cutoff α = 0.1. At higher stringencies, both filters eventually reduced rejections. For the ALL data, this effect occurred much more quickly for the overall mean filter. With the overall variance filter, the number of rejections increased by up to 50%. (D) Filtering on overall mean (θ = 0.5 is shown) removed many significant |Ti| (e.g., |Ti| > 4), while filtering on overall variance retained them.
), on the other hand, produced a small increase in rejections at low stringency, but then substantially reduced rejections, and thus power, at higher stringencies. (C) Effect of increasing filtering stringency for fixed adjusted p-value cutoff α = 0.1. At higher stringencies, both filters eventually reduced rejections. For the ALL data, this effect occurred much more quickly for the overall mean filter. With the overall variance filter, the number of rejections increased by up to 50%. (D) Filtering on overall mean (θ = 0.5 is shown) removed many significant |Ti| (e.g., |Ti| > 4), while filtering on overall variance retained them.