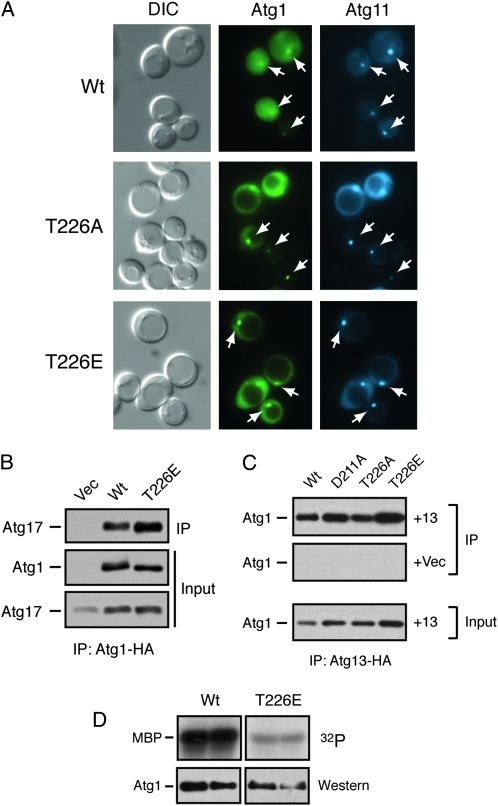
Figure 9.—
The Atg1T226E variant was localized to the PAS and interacted with Atg13 and Atg17. (A) The Atg1T226A and Atg1T226E proteins were localized normally to the PAS. The subcellular localization of YFP fusions to the indicated Atg1 proteins was assessed in an atg1Δ strain by fluorescence microscopy. An Atg11–CFP fusion protein was used as a marker for the PAS. The cells were all treated with 200 ng/ml rapamycin for 2 hr before examination. The white arrows indicate the location of the PAS in the respective cells. (B) The Atg1T226E variant interacted with Atg17. The indicated HA-tagged Atg1 proteins were precipitated from cell extracts and the relative amount of associated Atg17 protein was assessed by Western blotting with an anti-myc antibody. (C) The Atg1T226E variant interacted with Atg13. HA-tagged Atg13 was precipitated from cell extracts and the relative amounts of the associated Atg1 proteins were assessed by Western blotting with an anti-myc antibody. +13, cells containing the HA-tagged Atg13 protein; +Vec, cells containing a vector control and thus lacking the tagged Atg13 protein. (D) An in vitro kinase assay using MBP as substrate was performed with the wild-type Atg1 and the Atg1T226E variant. The indicated Atg1 proteins were immunoprecipitated and incubated with [γ-32P]ATP and the nonphysiological substrate, MBP; each sample was performed in duplicate. The bottom parts show the amount of Atg1 protein present in the in vitro reactions.