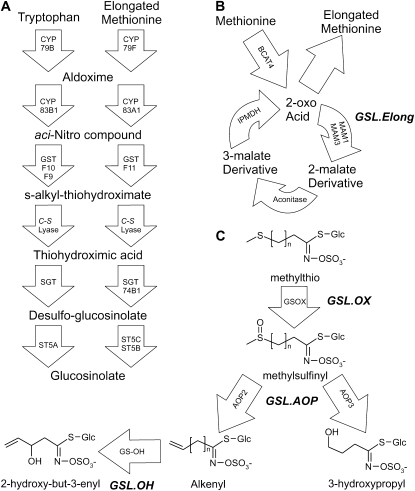
Figure 1.—
GSL biosynthesis and cloned QTL. Arrows show the known and predicted steps for GSL biosynthesis with the gene name for each biochemical reaction within the arrow. For undetected intermediate compounds, only chemical names are provided; for detected compounds, both the structure and chemical name are provided. The position of known genetic loci controlling biosynthetic variation is shown in bold italics. (A) The pathway and genes responsible for the production of the core GSL structure from tryptophan (indolic GSL) and methionine (aliphatic GSL). (B) Chain elongation cycle for aliphatic GSL production. Each reaction cycle adds a single carbon to a 2-oxo-acid, which is then transaminated to generate homomethionine for aliphatic GSL biosynthesis. The GSL.Elong QTL alters this cycle through variation at the MAM1, MAM2, and MAM3 genes that leads to differential GSL structure and content (Kroymann et al. 2001; Textor et al. 2004). (C) Aliphatic GSL side chain modification within the Bay-0 × Sha RIL population. Side-chain modification is controlled by variation at the GSL.ALK QTL via cis-eQTL at the AOP2 and AOP3 genes (Wentzell et al. 2007). The Cvi and Sha accessions express AOP2 to produce alkenyl GSL. In contrast, the Ler and Bay-0 accessions express AOP3 to produce hydroxyl GSL. Col-0 is null for both AOP2 and AOP3, producing only the precursor methylsulfinyl GSL (Kliebenstein et al. 2001a; Wentzell et al. 2007). The GSL.OX QTL appear to be controlled by cis-eQTL regulating flavin-monoxygenase enzymes (GS-OX1 to 5) that oxygenate a methylthio to methylsulfinyl GSL (Hansen et al. 2007; Li et al. 2008). The GSL.OH QTL is a cis-eQTL in the GS-OH gene, which encodes the enzyme for the oxygenation reaction (Hansen et al. 2008).