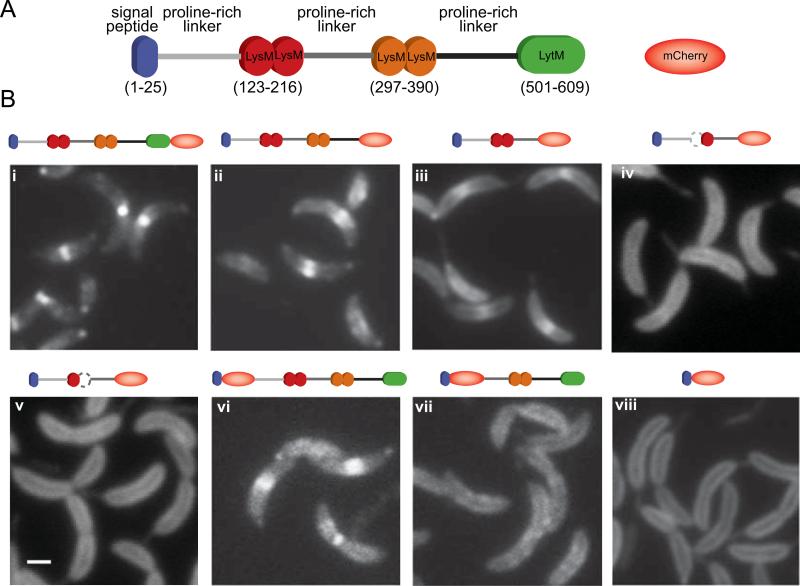
Fig. 4.
Determination of the DipM region involved in protein localization. (A) Schematic of the DipM domain organization and representation of mCherry. (B) Fluorescent micrographs of mCherry fused to different fragments of DipM: (i) DipM-mCherry (CJW3124); (ii) DipMΔ501-609-mCherry (CJW3439); (iii) DipMΔ297-609-mCherry (CJW3121); (iv) DipM1–296Δ121–167-mCherry (CJW3526); (v) DipM1–296Δ175–223-mCherry (CJW3528); (vi) mCherry-DipMΔ1-30 (CJW3116); (vii) mCherry-DipMΔ1-236 (CJW3117); (viii) DipMΔ54-609-mCherry (CJW2959). All inducible mCherry fusions were expressed from the chromosome by adding vanillic acid to a final concentration of 250 μM about 5 to 6 h prior to microscopy. In the case of the N-terminal mCherry fusions, the signal peptide is encoded by the vector. Note that wild-type DipM is also produced in these backgrounds. The white bar corresponds to 1 μm.