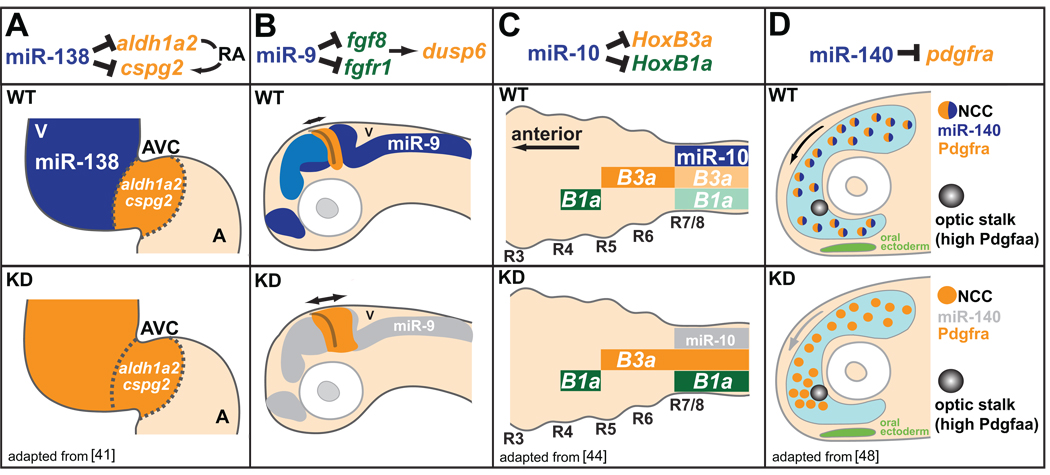
Figure 3.
miRNAs shape gene expression boundaries and migratory behaviors. (A) miR-138 (expression in blue) represses RA signaling in the ventricle (V) by targeting atrioventricular canal (AVC)-specific genes aldh1a2 and cspg2. miR-138 knockdown causes ventricular expression of AVC-specific genes (orange), and results in defects in ventricle morphology and function. A, atrium. (B) miR-9 expression (blue) adjacent to the mid-hindbrain boundary (MHB) delimits the extent of FGF signaling originating from the MHB. Knockdown of miR-9 leads to increased expression of FGF target genes, and the ectopic expansion of the FGF target dusp6 (orange). (C) miR-10 (expression in blue) dampens HoxB3a (orange) and HoxB1a (green) posterior expression in the nervous system. Knockdown of miR-10 results in increased expression of HoxB3a and HoxB1a within posterior domains. (D) miR-140 represses pdgfra expression within a subset of neural crest cells (NCCs) involved in palatogenesis to regulate migratory response to the Pdgfaa attractant (light blue). Knockdown of miR-140 results in increased expression of pdgfra, and defects in NCC migration around the optic stalk, a key source of Pdgfaa, leading to palate clefting.