Table 1.
Sequencing statistics of analyzed libraries.
| Ath (random) | Pam (normal) | Eca (oligo-dT) | Eca (random) | Eca (combined) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type | n | 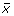 |
n | 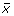 |
n | 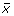 |
n | 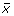 |
n | 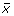 |
| Reads | 134,791 | 102.2 | 269,057 | 85.9 | 251,716 | 98.9 | 307,836 | 98.2 | 559,552 | 98.6 |
| Contigs | 8,188 | 147.0 | 22,303 | 107.3 | 18,339 | 148.5 | 14,242 | 146.9 | 30,603 | 159.1 |
| Singletons | 74,093 | 101.6 | 211,882 | 85.0 | 64,931 | 99.9 | 61,031 | 99.1 | 89,982 | 99.5 |
| Unigenes | 82,271 | 106.1 | 234,185 | 90.6 | 83,270 | 107.7 | 75,273 | 105.1 | 120,585 | 106.9 |
| MB | 13.8 | 23.1 | 24.9 | 30.2 | 55.1 | |||||
Read, Contig, Singleton, and Unigene Counts (n), mean sequence lengths (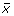 ), and total amount of sequence data (MB) for 454 GS20 libraries analyzed. Species codes are Ath (Arabidopsis thaliana), Pam (Persea americana, avocado), Eca (Eschscholzia californica, California poppy). cDNA library production method indicated in parentheses. Read lengths based on number of Q20 equivalent bases produced, after trimming and cleaning with the program seqclean http://compbio.dfci.harvard.edu/tgi/software/; normalized library original read mean length was 100.1 prior to trimming normalization adapter.
), and total amount of sequence data (MB) for 454 GS20 libraries analyzed. Species codes are Ath (Arabidopsis thaliana), Pam (Persea americana, avocado), Eca (Eschscholzia californica, California poppy). cDNA library production method indicated in parentheses. Read lengths based on number of Q20 equivalent bases produced, after trimming and cleaning with the program seqclean http://compbio.dfci.harvard.edu/tgi/software/; normalized library original read mean length was 100.1 prior to trimming normalization adapter.
