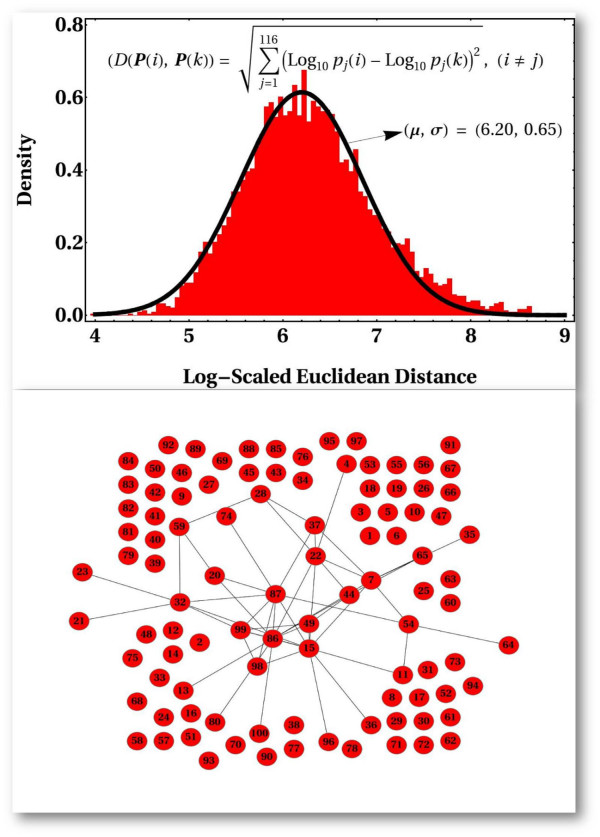
Figure 4.
Metric relations among reference dynamical trajectories distributed in different regions of biochemical reaction space. To more clearly appreaciate the possible metric relations among the 100 parameter configurations (reference points) distributed throughout biochemical reaction space that were selected, we calculated all possible distances (via the metric shown in the top panel) among configurations. We then fit the empirical distribution to a theoretical Normal distribution with parameters μ = 6.20 and σ = 0.65. With this information at hand, we constructed the graph shown in the bottom panel. This graph provides an interesting graphical notion of the possible metric relations among configurations in parameter space. We implemented a decision rule in order to construct the input adjacency matrix (a binary matrix) of the graph: if any element of the matrix A, aij, containing Log-scale Euclidean distances (see metric in top right panel) among parameter configurations is aij ⋜ μ - 2 * σ then aij → 1; otherwise aij → 0. The calculated graph is meant to illustrate how likely one point in parameter space (here represented by a node in the graph) can be accessed from another one via multiple perturbations. For example, pairs of linked nodes indicate that such configurations are relatively close in biochemical reaction space, and thus, one configuration might be accessed from the other via, perhaps, few random changes. In top right panel, P(i) and P(k) stand for any parameter configuration i or j included in the ensemble of trajectories analyzed.