Abstract
Background
Surface contamination of smear cheese by Listeria spp. is of major concern for the industry. Complex smear ecosystems have been shown to harbor antilisterial potential but the microorganisms and mechanisms involved in the inhibition mostly remain unclear, and are likely related to complex interactions than to production of single antimicrobial compounds. Bacterial biodiversity and population dynamics of complex smear ecosystems exhibiting antilisterial properties in situ were investigated by Temporal temperature gradient gel electrophoresis (TTGE), a culture independent technique, for two microbial consortia isolated from commercial Raclette type cheeses inoculated with defined commercial ripening cultures (F) or produced with an old-young smearing process (M).
Results
TTGE revealed nine bacterial species common to both F and M consortia, but consortium F exhibited a higher diversity than consortium M, with thirteen and ten species, respectively. Population dynamics were studied after application of the consortia on fresh-produced Raclette cheeses. TTGE analyses revealed a similar sequential development of the nine species common to both consortia. Beside common cheese surface bacteria (Staphylococcus equorum, Corynebacterium spp., Brevibacterium linens, Microbacterium gubbeenense, Agrococcus casei), the two consortia contained marine lactic acid bacteria (Alkalibacterium kapii, Marinilactibacillus psychrotolerans) that developed early in ripening (day 14 to 20), shortly after the growth of staphylococci (day 7). A decrease of Listeria counts was observed on cheese surface inoculated at day 7 with 0.1-1 × 102 CFU cm-2, when cheeses were smeared with consortium F or M. Listeria counts went below the detection limit of the method between day 14 and 28 and no subsequent regrowth was detected over 60 to 80 ripening days. In contrast, Listeria grew to high counts (105 CFU cm-2) on cheeses smeared with a defined surface culture.
Conclusions
This work reports the first population dynamics study of complex smear ecosystems exhibiting in situ antilisterial activity. TTGE revealed the presence of marine lactic acid bacteria that are likely related to the strong Listeria inhibition, as their early development in the smear occurred simultaneously with a decrease in Listeria cell count.
Background
The surface of traditional smear-ripened cheeses is colonized by a complex microbial ecosystem. Its biodiversity has been investigated by identification of cultivable isolates with molecular techniques, such as Pulsed-field gel electrophoresis (PFGE), Repetitive sequence-based PCR (rep-PCR) and 16S rDNA sequencing, or with Fourier-transform infrared spectroscopy (FTIR) [1-3]. Biodiversity studies using culture independent fingerprinting techniques, such as Temporal temperature gradient gel electrophoresis (TTGE), Denaturing gradient gel electrophoresis (DGGE), Single strand conformation polymorphism (SSCP) and Terminal restriction fragment length polymorphism (T-RFLP), have revealed the presence of additional uncultivable species [4-6]. The development of the smear is a dynamic process driven by metabiosis leading to the successive growth of several microbial communities. The first microorganisms to colonize the surface are yeasts. Yeasts' deacidification properties create a favorable environment for the next populations, mainly staphylococci followed by coryneforms. These two shifts in the microbial community structure of the smear have been observed in multiple studies [6-8]. Various marine bacteria have also been detected recently on cheese surface [5,9,10]. Population dynamics of complex cheese surface ecosystems at species level have been studied by cultivation methods, but these approaches are necessarily limited by the selectivity of the cultivation media chosen. Alternatively, fingerprinting techniques can be used to generate data on main populations of such ecosystems. These methods are fast and can give a more exhaustive view of the biodiversity in cheese but they are greatly influenced by the quality of DNA extraction protocols and bias may be introduced by the PCR amplification step [11]. The main advantage of TTGE/DGGE over other available fingerprinting techniques is the possibility to identify detected species by sequencing of DNA fragments after excision from the TTGE/DGGE gel [11]. TTGE/DGGE has been applied to study dominant bacteria of dairy products, enabling detection of species accounting for at least 1 to 10% of the total flora, depending on the amplification efficiency of the PCR step for a given species [4,12].
Surface contamination of smear cheese by Listeria monocytogenes is of concern for the industry since listeriosis breakouts have been associated with consumption of cheese [13]. Improvements in hygienic conditions and application of safety guidelines failed to reduce the contamination frequency to an acceptable level [14]. Growth of Listeria on cheese surface is closely linked to the development of the surface ecosystems and is primarily supported by yeast growth, which leads to deacidification and provides nutrients for bacterial growth. Listeria sp. has been shown to grow easily on smear cheeses when defined ripening cultures containing Debaryomyces hansenii, Geotrichum candidum and Brevibacterium linens were used [15,16]. Certain complex consortia naturally developing on smear cheese surface have been shown to inhibit Listeria sp. in situ [9,15,17]. In vitro studies of these anti-listerial activities led to the isolation of bacteriocin-producing strains among ripening microorganisms in certain cases [18,19]. Application of the bacteriocin producing strain on artificially contaminated cheeses failed however to fully restore the inhibition [15] or disturbed the development of the smear [20]. A better knowledge of microbial biodiversity and in situ population dynamics is crucial to identifying species that may be involved in the inhibition. Saubusse et al. [21] successfully used this approach for detecting antilisterial flora naturally developing in the core of Saint-Nectaire type cheese.
The objective of the present study was therefore to investigate population dynamics of complex cheese surface consortia with respect to their in situ inhibition properties. Two surface consortia were isolated from commercial Raclette type cheeses. TTGE was used for assessing biodiversity of both consortia at species level. An in-house database for species-level identification of the bands appearing in the TTGE fingerprints was developed with cultivable isolates. The two complex consortia or a control flora (defined commercial culture) were then applied on freshly-produced Raclette cheeses that were artificially contaminated with Listeria innocua. Population dynamics and Listeria growth were monitored over 60 to 80 ripening days.
Results
Bacterial biodiversity of cheese surface consortia by cultivation - Development of a TTGE profiles database
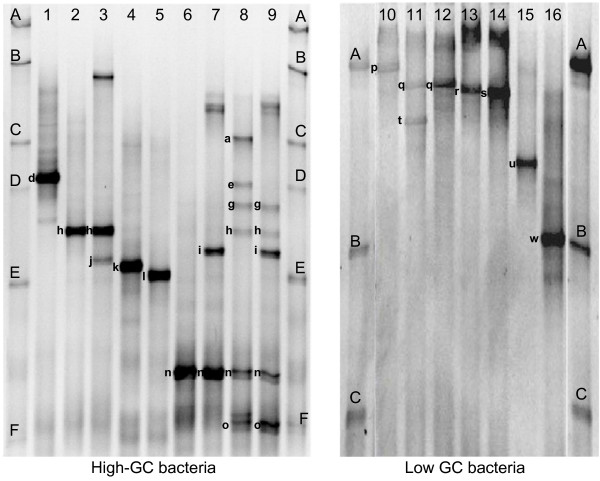
Consortium F was serial plated on five selective and non-selective media. A total of 128 cultivable isolates were subjected to TTGE fingerprinting analysis and grouped into 16 TTGE profiles. One representative isolate of each profile was randomly selected and subjected to 16S rDNA sequencing. Isolates displaying identical TTGE profiles but isolated from different cultivation media or exhibiting different macroscopic or microscopic morphologies were also subjected to 16S rDNA sequencing analyses. The presence of 15 species was detected by cultivation, with 7 dominant species enumerated on TGYA, the medium used for the determination of total cell count (Table 1). The number of bands and the corresponding migration lengths were recorded in a database (Figure 1). A majority of species displayed TTGE profiles with a single band for all isolates. Three species showed strain variations in TTGE profiles, with some strains harboring 1 to 5 supplementary bands (Figure 1). In addition, several species had indistinguishable TTGE profiles. Profile 5 corresponded to both Brachybacterium sp. and Arthrobacter arilaitensis, profile 12 to Staphylococcus equorum, Staphylococcus epidermidis and Facklamia tabacinasalis, and profile 16 to both Lactococcus lactis and Marinilactibacillus psychrotolerans (Figure 1). Low-GC bacteria Lc. lactis and M. psychrotolerans could not be distinguished on low-GC gel whereas high-GC gel revealed specific bands for the two species (bands z and z', respectively, in Figure 2). The database (Figure 1) contained a total of 16 TTGE profiles corresponding to 15 species. It was used as reference for species-level identification in TTGE fingerprints obtained by the culture independent approach.
Table 1.
Bacterial composition of cheese surface consortium F by a culture dependent method1
| Bacterial species | Accession number2 | Similarity (%) | Isolation media3 | Viable count [CFU cm-2] | Percentage on TGYA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Brevibacterium linens (or Brevibacterium aurantiacum4) |
GenBank:AJ315491 (GenBank:X765664) |
95.5-98.0 (97.8) |
TGYA | 7.5.108 | 32.5% | |||||
| Staphylococcus vitulinus | GenBank:NR_024670 | 99.6 | TGYA | 6.0.108 | 26.0% | |||||
| Brachybacterium tyrofermentans | GenBank:X91657 | 97.9 | TGYA | 4.5.108 | 19.5% | |||||
| Corynebacterium casei | GenBank:DQ361013 | 100.0 | TGYA | 1.5.108 | 6.5% | |||||
| Microbacterium gubbeenense | GenBank:AF263564 | 97.9 | TGYA | 1.5.108 | 6.5% | |||||
| Marinilactibacillus psychrotolerans | GenBank:AB083413 | 99.8 | TGYA | 1.5.108 | 6.5% | |||||
| Brachybacterium sp. | GenBank:AF513397 | 99.9 | TGYA | 0.7.108 | 3.0% | |||||
| Staphylococcus equorum | GenBank:NR_027520 | 98.8-99.1 | MSA | 3.0.108 | - | |||||
| Staphylococcus epidermidis | GenBank:NC_004461 | 98.5 | MSA | 8.107 | - | |||||
| Facklamia tabacinasalis | GenBank:Y17820 | 99.1 | BP | 6.105 | - | |||||
| Lactococcus lactis | GenBank:NC_002662 | 99.5 | MRS | 4.104 | - | |||||
| Enterococcus devriesei | GenBank:AJ891167 | 98.2 | MRS | 1.104 | - | |||||
| Enterococcus malodoratus | GenBank:Y18339 | 99.8 | MRS | 2.103 | - | |||||
| Enterococcus faecalis | GenBank:AJ420803 | 99.3 | KFS | 2.102 | - | |||||
| Enterococcus faecium | GenBank:EU547780 | 100.0 | KFS | 6.101 | - | |||||
1 128 isolates, i.e. ca. 25 isolates per media, were analyzed by TTGE and grouped into identical TTGE profiles. A representative isolate of each profile was identified by 16S rDNA sequencing. After the assignment of all isolates to a species, the percentage of each species on each of the five media was assessed. The cell count of a species was calculated by multiplying percentage and cell count determined for the corresponding media.
2 Closest 16S rDNA sequence in the GenBank public database http://www.ncbi.nlm.nih.gov.
3 Total cell count was determined on TGYA. In addition, staphylococci were enumerated on BP agar and MSA, lactic acid bacteria on MRS agar and enterococci on KFS agar.
4 Given the polymorphy in the intraspecies diversity of B. linens (Oberreuter et al. [52]), strain assignation to B. linens or the related species B. aurantiacum based on 16S rDNA analysis only was considered not reliable.
Figure 1.
Database for species-level identification of bands in TTGE fingerprints of complex cheese surface ecosystems. 128 isolates from consortium F were grouped into 16 TTGE profiles corresponding to 15 species. TTGE profiles 1-9 and 10-16 were analyzed on gels optimized for the separation of high-GC bacteria and low-GC bacteria, respectively. 1, Microbacterium gubbeenense (band d); 2, 3, Corynebacterium casei (bands h, j); 4, Brachybacterium tyrofermentans (band k); 5, Brachybacterium sp. or Arthrobacter arilaitensis from the ladder (band l); 6, 7, 8, 9, Brevibacterium linens (bands a, e, g, h, i, n, o); 10, Staphylococcus vitulinus (band p); 11, Staphylococcus equorum (bands q, t); 12, Staphylococcus equorum, Staphylococcus epidermidis or Facklamia tabacinasalis (band q); 13, Enterococcus malodoratus (band r); 14, Enterococcus faecium or Enterococcus devriesei (band s); 15, Enterococcus faecalis (band u); 16, Lactococcus lactis or Marinilactibacillus psychrotolerans (band w). Ladder: A, Lactobacillus plantarum SM71; B, Lactococcus lactis diacetylactis UL719; C, Corynebacterium variabile FAM17291; E, Arthrobacter arilaitensis FAM17250; D, F, Brevibacterium linens FAM17309.
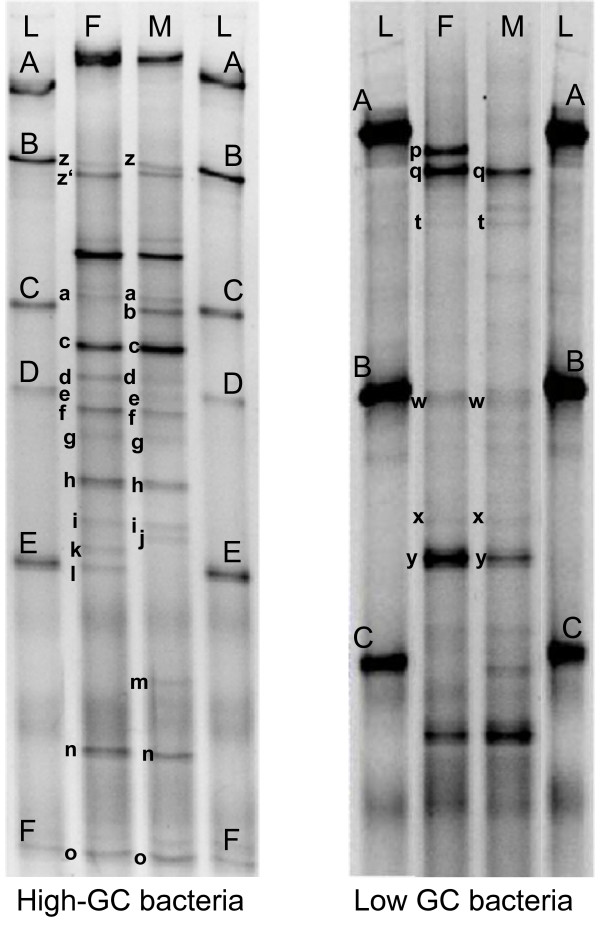
Figure 2.
Biodiversity of cheese surface consortia F and M by a culture independent method. TTGE fingerprints were analyzed on two different gels (high and low GC) after total DNA extraction of cheese surface consortia. Single bands were assigned to species using the species database or by excision, cloning and sequencing (*). b, c*, C. variabile; d, Mc. gubbeenense; f*, uncultured bacterium from marine sediment; h, j, C. casei; k, Br. tyrofermentans; l, Brachybacterium sp.; m*, Br. paraconglomeratum; a, e, g, h, i, n, o, B. linens; p, St. vitulinus; q, St. equorum, St. epidermidis or F. tabacinasalis; q, t, St. equorum; w, Lc. lactis or M. psychrotolerans; x*, Ag. casei; y*, Al. kapii; z, Lc. lactis; z', M. psychrotolerans. L, Ladder: A, Lb. plantarum SM71; B, Lc. lactis diacetylactis UL719; C, C. variabile FAM17291; E, A. arilaitensis FAM17250; D, F, B. linens FAM17309.
Bacterial biodiversity of cheese surface consortia by TTGE fingerprinting
Bacterial biodiversity of consortium F and M was assessed by TTGE fingerprinting of total DNA extracts, a culture independent method (Figure 2). Both consortia were analyzed on two gels, targeting the bacterial species with high-GC and low-GC content in separate runs. TTGE fingerprints of consortia F and M were composed of 18 bands each, corresponding to 13 and 10 species, respectively. Five bands could not be assigned to a known species of the database and were therefore submitted to cloning and sequencing after excision (Table 2). High similarity was found between consortium F and M with 9 common species, i.e. Corynebacterium variabile, Microbacterium gubbeenense, an uncultured bacterium from marine sediment (Table 2), Corynebacterium casei, Brevibacterium linens, Staphylococcus equorum, Lactococcus lactis, Agrococcus casei and Alkalibacterium kapii. Consortium F showed a higher diversity than consortium M with four additional species, Brachybacterium tyrofermentans, Brachybacterium sp., Marinilactibacillus psychrotolerans and Staphylococcus vitulinus. The species Brachybacterium paraconglomeratum was specific to consortium M.
Table 2.
Identification of non-assigned TTGE bands by excision, cloning and sequencing
| Band Designation1 | Bacterial species | Accession number2 | Similarity (%) |
|---|---|---|---|
| c | Corynebacterium variabile | GenBank:AJ783438 | 98.3 |
| f 3 | uncultured bacterium from marine sediment | GenBank:FJ717185 | 97.2 |
| m | Brachybacterium paraconglomeratum | GenBank:AJ415377 | 96.8 |
| x | Agrococcus casei | GenBank:DQ168427 | 100 |
| y | Alkalibacterium kapii | GenBank:AB294171 | 97.5 |
1 These designations are used to annotate bands from TTGE gels in figures 2 and 3.
2 Closest 16S rDNA sequence in the GenBank public database http://www.ncbi.nlm.nih.gov.
3The 16S rDNA sequence of band f exhibited highest similarity of 94% with Clostridiisalibacter paucivorans (GenBank: EF026082), a bacterium that belong to cluster XII of the Clostridium subphylum [53].
Population dynamics of cheese surface consortia by cultivation methods
Total cell counts and yeast counts were similar for all cheeses, independent of the surface flora applied to cheeses, i.e. consortium F, M or control flora OMK 704. Total cell counts increased from 1.2 ± 0.4 × 107 CFU cm-2 to 1.2 ± 0.7 × 109 CFU cm-2 within 14 days and remained stable afterwards (1.7 ± 1.0 × 109 CFU cm-2). Yeast counts increased from day 4 to reach 6.5 ± 0.2 × 106 CFU cm-2 at day 7 and decreased afterwards by 2 to 3 log until the end of ripening. Mould counts of ca. 102 CFU cm-2 were measured after 3 weeks ripening on cheeses treated with consortium F, while no moulds were detected on the cheese treated with consortium M or on control cheese. At the end of ripening, similar mould counts of ca. 104 CFU cm-2 were measured on all cheeses. The pH of cheese surface increased from 5.5 ± 0.1 at day 4 to 6.8 ± 0.4 at day 7 to 10, depending on the cheese, and was constant afterwards, with mean pH of 7.2 ± 0.4.
Population dynamics of complex cheese surface consortia by TTGE fingerprinting
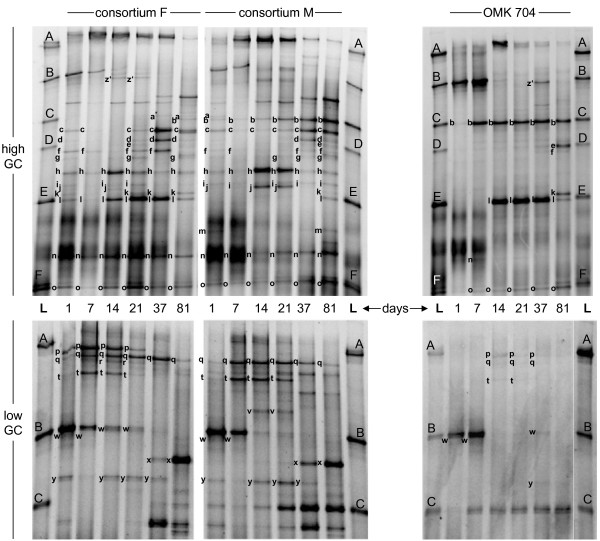
Population dynamics of consortium F or M were assessed at species level by TTGE fingerprinting of total DNA extracts (Figure 3, Table 3). TTGE fingerprints of day 1 cheese depict the starter culture (Lc. lactis) as well as the composition of the smear brines. Multiple shifts in the microbial community structure of cheeses treated with complex surface consortia F or M were observed throughout ripening. The nine species common to both consortia had similar sequential development on cheese surface. Lc. lactis, used as starter culture for cheese manufacture, was part of the dominant flora until day 7 and disappeared thereafter. St. equorum was the first species to colonize the surface within 7 days. Al. kapii grew on day 14 concomitant with C. casei and B. linens, followed by C. variabile, an uncultured bacterium from marine sediment and Mc. gubbeenense between day 14 and 37. Agrococcus casei was first detected on day 37. Other species specific to consortium F (St. vitulinus, Enterococcus sp., M. psychrotolerans, Brachybacterium sp. and Br. tyrofermentans) colonized the corresponding cheese after 7 to 21 days ripening. Both Brachybacterium species also colonized the cheese treated with consortium M, but could only be detected after 81 days, together with the Brachybacterium species specific to consortium M (Br. paraconglomeratum). Repetition of both treatments revealed the same trends with minor differences including a growth delay (ca. 5 days) for some high-GC species and the additional development of M. psychrotolerans at day 20 on the cheese treated with consortium M (data not shown).
Figure 3.
Population dynamics of cheese surface consortia by TTGE. TTGE analysis was carried out after total DNA extraction of cheese surfaces treated with complex surface consortium F, complex surface consortium M, or defined commercial culture OMK 704 (control cheese). Cheeses were sampled after 1, 7, 14, 21, 37 and 81 days. Each sample was analyzed on two different gels (high and low GC). Single bands were assigned to species using the database of 15 cultivable species completed by the database of 5 species identified by excision, cloning and sequencing. b, c, C. variabile; d, Mc. gubbeenense; f, uncultured bacterium from marine sediment; h, j, v, C. casei; k, Br. tyrofermentans; l, Brachybacterium sp. or Arthrobacter arilaitensis; m, Br. paraconglomeratum; a, e, g, h, i, n, o, B. linens; p, St. vitulinus; q, St. equorum, St. epidermidis or F. tabacinasalis; q, t, St. equorum; r, E. malodoratus; w, M. psychrotolerans or Lc. lactis; x, Ag. casei; y, Al. kapii; z', M. psychrotolerans. L, Ladder: A, Lb. plantarum SM71; B, Lc. lactis diacetylactis UL719; C, C. variabile FAM17291; E, A. arilaitensis FAM17250; D, F, B. linens FAM17309.
Table 3.
Population dynamics of cheese surface consortia by TTGE1
| Bacterial species detected with TTGE | Band designation2 | Consortium F (ripening day) | Consortium M (ripening day) | OMK 704 (ripening day) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 14 | 21 | 37 | 81 | 1 | 7 | 14 | 21 | 37 | 81 | 1 | 7 | 14 | 21 | 37 | 81 | |||||||
| Ag. Casei | x | + | d. | + | d. | |||||||||||||||||||
| Al. kapii | y | d. | + | d. | d. | d. | + | d. | d. | + | ||||||||||||||
| Br. paraconglomeratum | m | d. | + | |||||||||||||||||||||
| Brachybacterium sp., or A. arilaitensis | l | d. | d. | + | d. | d. | d. | + | + | d. | d. | d. | ||||||||||||
| Br. tyrofermentans | k | d. | + | d. | d. | + | + | |||||||||||||||||
| B. linens | a, e, g, h, i, n, o | d. | d. | + | d. | d. | d. | d. | d. | + | d. | d. | d. | + | d. | d. | d. | d. | ||||||
| C. casei | h, j, v | d. | d. | + | d. | d. | d. | d. | d. | + | d. | d. | d. | |||||||||||
| C. variabile | b, c | d. | d. | + | d. | d. | d. | d. | + | d. | d. | d. | d. | + | d. | d. | d. | d. | ||||||
| E. malodoratus | r | + | d. | |||||||||||||||||||||
| Lc. lactis | w (without z') | d. | d. | d. | d. | d. | d. | |||||||||||||||||
| M. psychrotolerans | w and z' | + | d. | + | ||||||||||||||||||||
| Mc. gubbeenense | d | d. | + | d. | d. | + | d. | |||||||||||||||||
| St. equorum, St. epidermidis, or F. tabacinasalis | q | d. | + | d. | d. | d. | d. | d. | + | d. | d. | d. | d. | + | d. | d. | ||||||||
| St. equorum | q and t | d. | + | d. | d. | d. | + | d. | d. | + | d. | |||||||||||||
| St. vitulinus | p | d. | + | d. | d. | + | d. | d. | ||||||||||||||||
| uncultured bacterium from marine sediment | f | d. | d. | + | d. | d. | d. | + | d. | + | ||||||||||||||
1 The letter (d.) indicates sampling times where a given species was detected in the TTGE gel. The symbol (+) indicates growth of a species in the smear. Growth was assumed in two cases, i.e. when a band was detected at a sampling time, while absent from the previous sampling time, or when the intensity of the detected band increased, compared to the previous sampling time (valid only at day 7 and 14, i.e. for a 1 log-increase of the total cell count).
2The same letter code as for band designation in Figure 3 was used.
Population dynamics of the defined commercial culture OMK 704 by TTGE fingerprinting
Population dynamics of the defined commercial culture OMK 704 at species level was assessed by TTGE fingerprinting of total DNA extracts (Figure 3, Table 3). All three species of the culture OMK 704 (C. variabile, A. arilaitensis and B. linens) established themselves on cheese surface during the first 14 days. Each of the five B. linens strains of the culture OMK 704 exhibited a distinguishable strain-specific TTGE profile (data not shown). The profile of B. linens FAM17309 (Bands e, o; Figure 3) was detected in the TTGE fingerprint of day 81 cheese, showing that this strain predominated over other B. linens strains at the end of ripening. Additional species not deliberately applied on the cheese colonized the cheese surface along ripening. Two staphylococci species (St. vitulinus; St. equorum) appeared on day 14 as well as M. psychrotolerans and Al. kapii on day 37. Br. tyrofermentans and an uncultured bacterium from marine sediment completed the high GC community at day 81. Repetition of the treatment revealed the same trends regarding the three defined species. However, the development of non-deliberately applied species was different in the repetition. Three additional species colonized the cheese, i.e. Enterococcus sp., C. casei, Ag. casei, while Br. tyrofermentans could not be detected (data not shown).
In situ inhibition of Listeria by complex surface consortia
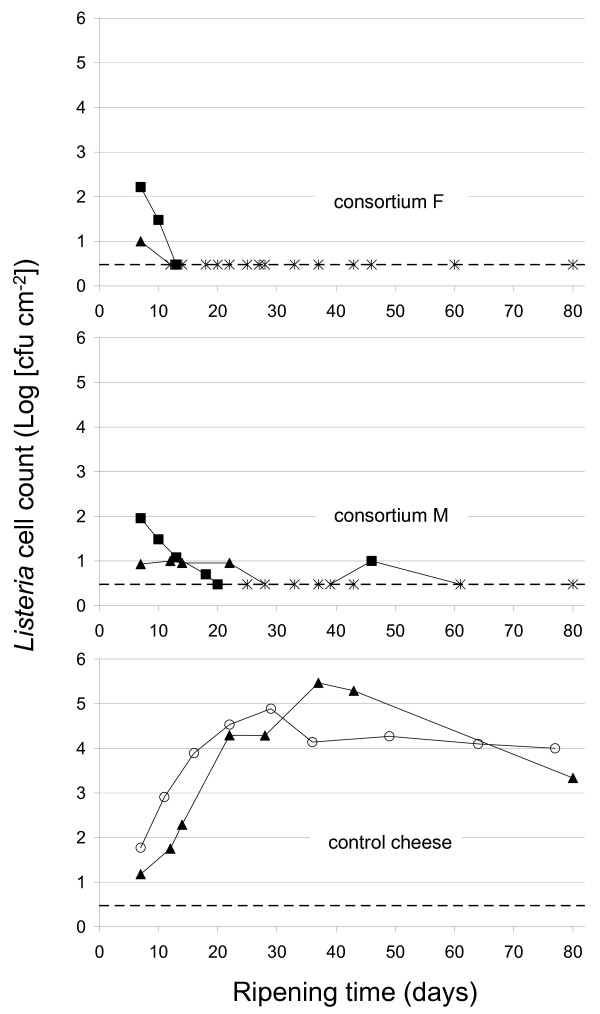
Raclette cheeses were artificially contaminated with 4 strains of Listeria innocua on day 7 and 8, i.e. when yeasts on cheese surface had reached high counts of 6.5 ± 0.2 × 106 CFU cm-2. From the amount added to the smear brine (5 × 103 CFU ml-1), Listeria counts of 1.4 ± 0.9 × 101 CFU cm-2 (first trial) and of 1.0 ± 0.6 × 102 CFU cm-2 (repetition) were recovered from the surface immediately after contamination. Listeria development was strongly affected by the surface flora applied for ripening. A decrease of Listeria counts below the detection limit of the method (< 3 CFU cm-2) was observed for cheeses treated with complex consortia F or M supplemented with Debaryomyces hansenii FAM14334 (Figure 4). Listeria could be recovered from cheese surface (~2000 cm2) with an enrichment procedure at the end of ripening (60 to 80 days), for both consortia. In contrast, Listeria counts on control cheeses treated with the commercial culture OMK 704 increased to ca. 105 CFU cm-2 after one month (Figure 4).
Figure 4.
In situ inhibition of Listeria on cheese surface by complex consortia. Cheese surfaces were treated with smear brines (3.3% (w/v) NaCl), inoculated with either consortium F, consortium M or the defined commercial culture OMK 704 (control cheese). Two independent experiments were carried out for each treatment. Different symbols indicate different commercial cheese production. Smear brines were inoculated with Listeria innocua on day 7 and 8, at 5 × 103 CFU ml-1. Stars indicate times where Listeria counts were below the detection limit of the enumeration method (< 3 CFU cm-2; dashed line).
Discussion
To our knowledge, this work describes the first dynamic study of naturally developing anti-listerial cheese surface consortia. The monitoring of two complex consortia obtained from industrial productions was carried out with TTGE, a culture independent fingerprinting technique which enabled species-level detection of high-GC and low-GC bacteria in separate runs.
Previous studies reported a broad range of biodiversity in smear consortia, with 2 to 15 bacterial species detected [2,5,22,23]. High bacterial diversity was observed in consortium F, with 13 species detected at dominant level by culture independent analysis. The cultivation approach detected only 9 of the 13 species present at dominant level in consortium F, but enabled detection of 6 additional species present at subdominant level. TTGE is a semiquantitative approach with limited sensitivity compared to the cultivation approach. However, as fingerprinting technique, TTGE enabled to overcome the arbitrary selection exercised on the flora by the cultivation step, giving a more complete view of biodiversity at dominant level. The combined use of both approaches led to a detailed knowledge of biodiversity in cheese smear flora, as already observed by Feurer et al. and Mounier et al. [5,24]. The identification strategy used in the present study for the cultivation approach, i.e. all cultivable isolates grouped by TTGE profiles and subsequent sequencing, enabled the detection of intraspecies diversity differentiation in 3 dominant species. This strategy greatly simplified the identification of bands in the TTGE fingerprints of complex consortia corresponding to intraspecies variability. Consortium M displayed slightly less diversity than F with 10 species detected at the dominant level by culture independent analysis.
A total of 20 species were detected in consortia F and M, including eight coryneform bacteria. C. variabile, C. casei, B. linens and Mc. gubbeenense are common ripening microorganisms of smear cheeses detected on soft cheeses [5,9] and semi-hard cheeses [2,8,23]. Br. tyrofermentans was first isolated from Gruyère cheese [25] and was recently shown to colonize the surface of soft cheeses [5,9]. To our knowledge, this is the first time that Br. paraconglomeratum has been detected in cheese although this species has been previously isolated from milk [26]. Agrococcus casei was first isolated from Gubbeen, an Irish semi-hard cheese [2]. Three Staphylococcus species were isolated in addition to coryneforms. St. equorum is common on smear cheeses [6,8,27-29] while St. vitulinus was only isolated by Irlinger et al. [27] from French cheeses. St. epidermidis, a human skin inhabitant, was detected on various Irish semi-hard cheeses [2,8]. Two Gram-positive marine lactic acid bacteria (LAB) and an uncultured bacterium from marine sediment were also part of the dominant flora. M. psychrotolerans has been detected in the smear of soft cheeses from Germany and France [5,9]. Alkalibacterium sp. was found to be present in many European cheeses including Tilsiter, a semi-hard smear cheese [10]. We also identified potentially undesirable species of enterococci in the subdominant flora of consortium F. Enterococci have a controversial status in the dairy industry. They are considered naturally occurring ripening organisms for artisan Mediterranean cheese [30], but also appear as emerging pathogens due to the virulence factors they tend to harbor [31]. To our knowledge, this study is the first report of the presence of Facklamia sp. in cheese. F. tabacinasalis was first isolated from powdered tobacco by Collins et al. [32] and has recently been detected in raw milk by Delbès et al. [33] in a French farm producing Saint-Nectaire cheese and by Hantsis-Zacharov and Halpern [34] in a farm from northern Israel equipped with modern automated milking facilities. The presence of F. tabacinasalis on the surface of smear cheese may constitute a health hazard, as this species was shown to be α-haemolytic on horse blood [32]. Moreover, from six Facklamia species described to date, four were isolated from human clinical specimen [35].
We observed highly similar microbial community structures of consortia F and M, with 9 species being common to both consortia at dominant level, despite different ripening procedures. High interbatch diversity was described by Rea et al. [8] in a single cheese ripening facility of Gubbeen, an Irish semi-hard smear cheese, over 8 years production, which may be related to a lack of humidity and temperature control during ripening of Gubbeen cheese. In the present study, the production of Swiss Raclette type cheese with defined production and ripening parameters led to the development of a similar flora in two distinct dairies. The source of this highly diverse flora remains unidentified but possible sources could be the brine bath, skin of the workers or wooden shelves, as shown by Mounier et al. [36] for Gubbeen cheese.
The high biodiversity is particularly surprising in the case of dairy F, where the smear brine is freshly prepared prior to each smearing and inoculated with a defined ripening culture of only 3 bacterial species. Moreover, the smear brine is applied by a cheese ripening robot that smears the young cheeses first. However, the microflora of the brine bath is not controlled and might be one of the major sources. In particular, the brine bath (18-22% (w/v) NaCl) could be suitable to maintain the two halophilic and alkaliphilic marine LAB detected in consortium F, as some strains of M. psychrotolerans and Al. kapii were shown to grow at salt concentration as high as 21% (w/v) by Ishikawa et al. [37,38].
Dynamic studies of consortia F and M inoculated at same cell counts on cheese surface revealed a similar sequential development of nine bacterial species, i.e. Lc. lactis, St. equorum, Al. kapii, C. casei, B. linens, C. variabile, an uncultured bacterium from marine sediment, Mc. gubbeenense and Ag. casei. The development of this microbial community prevented growth of Listeria innocua, inoculated at 5 × 103 CFU ml-1 smear brine on cheeses at day 7 and 8, over 60 to 80 days ripening. Contamination at day 7 and 8, i.e. when yeasts reached their highest density, provided optimal growth conditions for Listeria, as shown by the rapid Listeria growth on control cheese. Strong antilisterial activities were shown in this unfavorable condition for consortia F and M. Antilisterial activities of complex undefined cheese surface consortia were already observed in previous studies [9,15]. Maoz et al. [9] reported a total inhibition of L. monocytogenens during 40 days ripening of a soft smear cheese with an initial contamination level of 1.6 × 103 CFU ml-1 smear brine.
The surface of smear cheese contains a limited range of substrates supporting growth of microorganisms, mainly lactose and lactate. Lactose is mostly metabolized by LAB during curd acidification and initial ripening. The residual lactose can be metabolized on the cheese surface by yeasts during the first days of ripening, as shown for soft cheeses by Leclercq-Perlat et al. [39]. Lactate metabolized by yeasts into CO2 and H2O leads to deacidification of the cheese surface [40]. As a result, lactate continuously diffuses from the core to the surface of the cheese. Lactate can be totally consumed by surface microorganisms in soft cheeses [41]. Several smear bacterial species, i.e. Brevibacterium aurantiacum, C. casei, C. variabile, Mc. gubbeenense and St. saprophyticus, were shown to use lactate and casaminoacids for growth [42]. In contrast, Listeria sp. can only use a limited range of carbon sources for growth, including glucose, glycerol, fructose and mannose, while no growth occurs on lactate or casaminoacids [43-46]. Premaratne et al. [44] showed that Listeria monocytogenes may utilize alternative carbon sources, such as N-acetylglucosamine and N-acetylmuramic acid, which are major components of bacterial and fungal cell walls [44,47]. In addition, the yeast cell wall contains a mannan glycopeptide with mannose [48], a sugar metabolized by Listeria sp. Listeria growth on smear cheese can therefore be limited by a low availability of carbon source and stimulated by components of smear microorganisms.
Marine LAB ferment glucose into lactate and assimilate mannose [37,38]. Ishikawa et al. [38] reported that Al. kapii can utilize a fairly limited range of carbon sources. In the present study, M. psychrotolerans and/or Al. kapii established early on cheeses treated by complex consortia, i.e. between day 14 and day 20. We believe competition for nutrients between marine LAB and Listeria sp. may be involved in Listeria inhibition in the smear since the development of M. psychrotolerans and Al. kapii occurred simultaneously with the decrease of Listeria counts for both cheeses treated with consortium F (first trial and repetition) and for one cheese treated with consortium M (repetition). In addition, Listeria growth on control cheese stopped when M. psychrotolerans and Al. kapii were first detected in the smear, i.e. on day 37. Hain et al. [49] reported a microarray experiment conducted with the antilisterial complex smear consortium described by Maoz et al. [9]. Genes involved in energy supply were mostly up-regulated after 4 hours of contact between Listeria monocytogenes and the consortium, suggesting that Listeria had entered a state of starvation. While Maoz et al. [9] detected M. psychrotolerans in the aforesaid smear consortium by cultivation methods, they may have overlooked the presence of Al. kapii or related species.
Conclusions
This work reports the first study of population dynamics of antilisterial cheese surface consortia. Dynamics of two consortia obtained from industrial productions revealed highly similar, with the sequential development of 9 common species, whereas development of both consortia inhibited Listeria growth over the whole ripening period. Next to common cheese surface bacteria, the two consortia contained marine lactic acid bacteria (LAB) that developed early in ripening, shortly after the growth of staphylococci and concomitant with a decrease in Listeria cell counts. Competition for nutrients between marine LAB and Listeria sp. could be involved in the observed inhibition. Temporal temperature gradient gel electrophoresis revealed decisive to detect all marine bacteria present at dominant level in the smear, as only one of three species was detected by the culture dependent approach. Further cheese ripening experiments are needed to investigate the potential contribution of marine LAB to antilisterial activity.
Methods
Collection of cheese surface consortia and microbial cultures
Cheese surface consortia were collected from two Swiss cheese manufacturers of Raclette type cheese made of pasteurized milk. Consortium F was collected from a 4-weeks old cheese produced with a defined surface ripening culture in industrial-scale dairy F. The surface ripening culture was composed of OFR9 (Danisco A/S, Copenhagen, Denmark), containing Brevibacterium linens, Brevibacterium casei as well as three yeasts, and OMK 703 (Research Station Agroscope Liebefeld-Posieux ALP, Bern, Switzerland), containing Brevibacterium linens, Arthrobacter arilaitensis as well as two yeasts. Consortium M was collected from a 6-weeks old cheese in small-scale dairy M, where the cheeses were treated with old-young smearing, with a smear brine derived from Gruyère type cheese. Surface consortia were scraped off the cheese (~2000 cm2; ~10 g), homogenized in a stomacher in 100 ml 3.3% (w/v) NaCl for 4 min and stored at 4°C until further use but not longer than 30 days. Long-term storage (up to 7 months) was carried out by addition of 20% glycerol and freezing at -30°C. The commercial surface culture OMK 704 (ALP, Bern, Switzerland), used as control in cheese ripening experiments, contained one yeast (Debaryomyces hansenii FAM14334, ALP culture collection), five Brevibacterium linens, five Corynebacterium variabile, and one Arthrobacter arilaitensis strains. Each strain of the commercial culture was provided in a liquid form and stored at 4°C (short term) or at -30°C with addition of 20% glycerol (long term). For safety reasons, non pathogenic Listeria strains were used as a model for L. monocytogenes in cheese ripening experiments. Listeria innocua 80945-8, 81000-1, 81003-3, and 81587-4 (Laboratory of Food Biotechnology, ETH Zurich, Zurich, Switzerland) had previously been isolated from smears by ALP (Bern, Switzerland). Listeria strains were grown in tryptic soy broth (Oxoid, Pratteln, Switzerland) supplemented with 0.6% (w/v) yeast extract (Merck, Dietikon, Switzerland) at 30°C for 14 h.
Cell enumeration of cheese surface consortia
Total cell counts were determined on TGYA (Tryptic Glucose Yeast Agar, Biolife, Milano, Italy) supplemented with 1% (w/v) casein peptone (BBL, Heidelberg, Germany) after incubation at 30°C for 3 days, followed by incubation at room temperature with daylight exposure for another 7 days. Staphylococci were counted on BP agar (Baird Parker RPF agar; BioMérieux, Geneva, Switzerland) and MSA (Mannitol Salt Agar, Oxoid, Pratteln, Switzerland) after incubation at 37°C for 6 days. Yeast counts and mould counts were both determined on PY agar (Phytone Yeast extract agar, BBL, Heidelberg, Germany) incubated at 30°C. The plates were examined after 3 days for yeasts and after 6 days for moulds. Enterococci were determined on KFS agar (KF Streptococcus agar, Becton Dickinson AG, Allschwil, Switzerland) incubated at 42°C for 3 days, and Listeria on Palcam agar (Oxoid, Pratteln, Switzerland) incubated at 37°C for 2 days, all under aerobic conditions. Lactic acid bacteria were counted on MRS agar with Tween 80 (De Man et al., 1960, Biolife, Milano, Italy) incubated at 37°C for 6 days, under anaerobic conditions which were generated using GENbox anaerobic systems (Biomérieux, Geneva, Switzerland). At the end of ripening, the presence or absence of Listeria was assessed using a three-step enrichment procedure that was previously validated against the reference method ISO 11290-1 for use on smear samples by ALP (Bern, Switzerland). 10 g (~2000 cm2) of smear were homogenized in 90 g tryptic soy broth supplemented with 0.6% (w/v) yeast extract, 0.02% (w/v) Delvocid® (DSM, Heerlen, Netherlands), 0.001% (w/v) acriflavin (Fluka, Buchs, Switzerland), and 0.004% (w/v) nalidixic acid (Fluka, Buchs, Switzerland) for 4 min using a Stomacher and incubated at 30°C for 24 h. After this step, 1% (v/v) of enriched sample was inoculated to supplemented tryptic soy broth and incubated again at 30°C for 24 h. Presence or absence of Listeria was then checked by streaking a loopful of the second enrichment media on ALOA agar (Biolife, Pero, Italy) that was incubated at 37°C for 24 h.
DNA extraction of complex consortia and single isolates
Total DNA extraction of cheese surface consortia was carried out with 1 ml homogenate containing 107 to 109 CFU ml-1 that was centrifuged at 18'000 × g for 5 min. The resulting pellet was stored at -20°C until further use. The DNA extraction protocol was modified from Chavagnat et al. [50]. The frozen pellet was resuspended in 1 ml 0.1 M NaOH, incubated at room temperature for 15 min and centrifuged at 18'000 × g for 5 min. The pellet was resuspended in 1 ml TES buffer (10 mM EDTA, 0.1. M tris(hydroxymethyl)-aminomethane, 25% (w/v) saccharose) containing 0.25% (w/v) lysozyme (50000 U mg-1, Merck, Dietikon, Switzerland), incubated at 37°C for 1 h, and centrifuged at 18'000 × g for 5 min. The pellet was resuspended in 190 μl G2 Buffer (EZ1 DNA Tissue Kit, Qiagen, Basel, Switzerland) and 10 μl proteinase K (EZ1 DNA Tissue Kit; Qiagen, Basel, Switzerland) were added. This suspension was incubated at 56°C for 1 h after which DNA was further purified by BioRobot® EZ1 (Qiagen, Basel, Switzerland) and analyzed by TTGE, as described below.
DNA extraction of single isolates was carried out by dissolving one colony of a pure culture in 0.2 ml tris-K buffer (0.01 M tris(hydroxymethyl)-aminomethane (Merck, Dietikon, Switzerland)) containing 0.5 μl ml-1 Tween 20 (Fluka, Buchs, Switzerland) and 0.24 mg ml-1 proteinase K (Sigma-Aldrich, St. Louis, USA). This suspension was first heated at 60°C for 1 h followed by 95°C for 15 min, and centrifuged at 10'000 × g for 5 min resulting in a DNA containing supernatant that was further analyzed by TTGE.
Temporal temperature gradient gel electrophoresis (TTGE)
PCR amplification of the V3 region of the 16S rDNA (~200 bp) was performed according to Ogier et al. [12] using a Biometra T-Personal thermocycler (Biometra, Göttingen, Germany) with direct amplification using primers HDA1-GC and HDA2 (Microsynth, Balgach, Switzerland) and ~100 ng of bacterial DNA. Ten μl of PCR products were separated on a 2% (w/v) agarose gel to check successful amplification with a molecular weight standard of TriDye 100 bp DNA Ladder (BioConcept, Allschwil, Switzerland). TTGE analysis was carried out as described by Ogier et al. [12] with the following modifications. The electrophoresis was run in 1.5 × TAE buffer (1.5 mM EDTA, 60 mM tris(hydroxymethyl)-aminomethane, 60 mM acetic acid) at 65 V for 16 h, with a temperature ramp of 0.3°C h-1 from 66 to 70°C. The gel concentrations were optimized to enable visualization in separate runs of high-GC bacteria (8 M urea; 8.5% (w/v) acrylamide (37.5:1)) and low-GC bacteria (7 M urea; 8% (w/v) acrylamide (37.5:1)) by empirical approach using a ladder of dairy bacteria harboring a wide range of GC-contents (from 49% for Lactobacillus plantarum to 60% for Propionibacterium sp.). Volumes of 20 μl (isolates) or 30 μl (complex consortia) of PCR products were mixed with 20 μl loading dye (0.25% (w/v) Orange G, 50% (w/v) sucrose; Fluka, Buchs, Switzerland) and loaded in each well. The detection limit of the method proved similar to Ogier et al. [12], with detection of bacterial species accounting for at least 1% of the total DNA amount.
Identification of single isolates by partial sequencing of 16S rDNA
Groups of isolates with identical TTGE profiles were formed and a representative isolate of each group was selected for further 16S rDNA sequencing analyses. A 1400-bp fragment of the 16S rDNA was amplified with universal primers 16SUNI-L and 16SUNI-R [51]. The 50-μl reaction mixture contained ~20 ng DNA (NanoDrop® ND-100, Witec AG, Littau, Switzerland), 2.5 U of Taq DNA polymerase (Euroclone, Pero, Italy), 0.4 μM of each primer (Microsynth, Balgach, Switzerland), 200 μM of each deoxynucleoside triphosphate (Amersham Biosciences, Otelfingen, Switzerland), and the reaction buffer (Euroclone, Pero, Italy) consisting of 10 mM Tris-HCl, 50 mM KCl, and 1.5 mM MgCl2. The amplification was performed in a Biometra T-Personal thermocycler (Biometra, Göttingen, Germany) with the following temperature profile: 94°C for 3 min, 35 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 60 s, and a final annealing at 72°C for 7 min. Amplified DNA was purified using the GFX-PCR DNA Purification Kit (GE Healthcare Biosciences, Otelfingen, Switzerland). Partial sequencing was carried out with primer 16SUNI-L and the BigDye® Terminator v1.1 cycle sequencing kit (Applied Biosystems, Rotkreuz, Switzerland) and analyzed in an ABI Prism 310 genetic analyzer (Applied Biosystems, Rotkreuz, Switzerland). Species identification was obtained by matching the obtained partial sequence (500 to 900 bp) to deposited sequences in the GenBank public database using the BLAST program.
Identification of TTGE bands by partial sequencing of the 16S rDNA
Bands of the complex TTGE fingerprints that could not be identified by comparison with the database were excised, cloned and sequenced as described by Ogier et al. [12]. The eluted DNA was amplified by PCR using primers HDA1 and HDA2 (Microsynth, Balgach, Switzerland). PCR products were purified using the GFX-PCR DNA Purification Kit (GE Healthcare Biosciences, Otelfingen, Switzerland), ligated into pGEM®-T Easy vector (Promega, Dübendorf, Switzerland) and transformed into Escherichia coli (Subcloning Efficiency™ DH5™ Competent Cells, Invitrogen, Basel, Switzerland). After plasmid purification, the insert was amplified by PCR with primers HDA1-GC and HDA2. The PCR product was analyzed by TTGE to confirm its position in the gel and sequenced from both sides with primers HDA1 and HDA2. The sequence obtained (~200 bp) was matched to deposited sequences in the GenBank public database.
Cheese ripening experiments
Raclette type cheeses (~6 kg; 2000 cm2) produced from pasteurized milk in dairy F were taken immediately after brining. A water content of 44.9% (w/w) and salt content of 1.8% (w/w) were measured in a 24 h-old cheese from the production batch, by gravimetric analysis (ISO 5534/IDF 4:2004) and by potentiometric titration (IDF Standard 88A:1988), respectively. Cheeses were ripened in a pilot plant cheese cellar with controlled temperature at 11°C and relative humidity at 95% for 2 to 3 months. Cheeses were smeared daily until day 15 and twice a week thereafter, using 20 ml smear brine (3.3% (w/v) NaCl) per cheese side. Three different treatments were applied on cheeses and two independent experiments were carried out for each treatment. Cheeses were treated with 20 ml of smear brines inoculated with 5 × 108 CFU ml-1 of either: consortium F, consortium M or the commercial culture OMK 704. In addition, 1 × 107 CFU ml-1 of the yeast strain Debaryomyces hansenii FAM14334 were inoculated in all smear brines. Smear brines were prepared fresh before each smearing with the following protocol. The appropriate amounts of consortium or defined culture and yeast were added in a 50 ml centrifugation tube and the volume was adjusted to 20 ml by addition of 3.3% (w/v) NaCl. Tubes were then centrifuged at 5'000 × g for 15 min, and the pellet was resuspended in 20 ml of fresh 3.3% (w/v) NaCl.
Cheeses were artificially contaminated twice with Listeria after 7 and 8 days ripening. Listeria inoculum was prepared as follows. Overnight cultures of 4 Listeria innocua strains were mixed in a 1:1:1:1 ratio, diluted 10'000 times in 0.9% (w/v) NaCl, and 0.3 ml of the dilution were added to each smear brine after the centrifugation step, to reach a concentration of ca. 5 × 103 CFU ml-1. The brushes and wooden shelves were washed and autoclaved prior to each use. Wooden shelves were first changed after one week and every three weeks thereafter. The pH of the cheese surface was periodically measured in situ using a flat membrane electrode (InLab® Surface, Mettler-Toledo, Greifensee, Switzerland).
Microbial analyses of cheese surface during ripening experiments
Approximately 25 cm2 of cheese surface were scraped off using sterile cotton rolls (IVF Hartmann, Neuhausen, Switzerland) and aseptically transferred into a stomacher bag. Each sample was suspended in 25 ml pre-heated (45°C) peptone water, composed of 1% (w/v) casein peptone, 0.5% (w/v) NaCl and 2% (w/v) tri-sodium citrate dehydrate, all from Merck (Dietikon, Switzerland), and homogenized for 4 min using a Stomacher (Silver Masticator; IUL Instruments GmbH, Königswinter, Germany). 1 ml of this solution was submitted to total DNA extraction for TTGE as described above. Serial dilutions in 0.9% (w/v) NaCl were prepared and plated on TGYA, PY agar and Palcam agar. At the end of ripening, 10 g of smear were harvested and tested for the presence of Listeria using an enrichment procedure as described above.
Authors' contributions
ER carried out the experiments, evaluated the results and drafted the manuscript. MH participated in the creation of the TTGE database and in the repetition of the cheese experiment. SMS and EEM participated in the conception and coordination of the study and revision of the manuscript. CL provided guidance during the whole study and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Emmanuelle Roth, Email: emmanuelle.roth@alp.admin.ch.
Susanne Miescher Schwenninger, Email: susanne.miescher@ilw.agrl.ethz.ch.
Madlen Hasler, Email: mahasler@student.ethz.ch.
Elisabeth Eugster-Meier, Email: elisabeth.eugster@alp.admin.ch.
Christophe Lacroix, Email: christophe.lacroix@ilw.agrl.ethz.ch.
Acknowledgements
This work was supported by the Research Station Agroscope Liebefeld-Posieux ALP, Bern, Switzerland. We thank Daniel Goy for sharing expertise in cheese ripening. We also thank Hélène Berthoud and Monika Haueter for excellent assistance with sequencing and DNA extraction protocols.
References
- Bockelmann W, Hoppe-Seyler T. The surface flora of bacterial smear-ripened cheeses from cow's and goat's milk. Int Dairy J. 2001;11:307–314. doi: 10.1016/S0958-6946(01)00060-7. [DOI] [Google Scholar]
- Mounier J, Gelsomino R, Goerges S, Vancanneyt M, Vandemeulebroecke K, Hoste B, Scherer S, Swings J, Fitzgerald GF, Cogan TM. Surface microflora of four smear-ripened cheeses. Appl Environ Microbiol. 2005;71:6489–6500. doi: 10.1128/AEM.71.11.6489-6500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenning M, Theilmann V, Scherer S. Rapid analysis of two food-borne microbial communities at the species level by Fourier-transform infrared microspectroscopy. Environ Microbiol. 2006;8:848–857. doi: 10.1111/j.1462-2920.2005.00971.x. [DOI] [PubMed] [Google Scholar]
- Ogier JC, Lafarge V, Girard V, Rault A, Maladen V, Gruss A, Leveau JY, Delacroix-Buchet A. Molecular fingerprinting of dairy microbial ecosystems by use of temporal temperature and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2004;70:5628–5643. doi: 10.1128/AEM.70.9.5628-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurer C, Irlinger F, Spinnler HE, Glaser P, Vallaeys T. Assessment of the rind microbial diversity in a farmhouse-produced vs a pasteurized industrially produced soft red-smear cheese using both cultivation and rDNA-based methods. J Appl Microbiol. 2004;97:546–556. doi: 10.1111/j.1365-2672.2004.02333.x. [DOI] [PubMed] [Google Scholar]
- Rademaker JLW, Peinhopf M, Rijnen L, Bockelmann W, Noordman WH. The surface microflora dynamics of bacterial smear-ripened Tilsit cheese determined by T-RFLP DNA population fingerprint analysis. Int Dairy J. 2005;15:785–794. doi: 10.1016/j.idairyj.2004.08.027. [DOI] [Google Scholar]
- Bockelmann W. Development of defined surface starter cultures for the ripening of smear cheeses. Int Dairy J. 2002;12:123–131. doi: 10.1016/S0958-6946(01)00152-2. [DOI] [Google Scholar]
- Rea MC, Görges S, Gelsomino R, Brennan NM, Mounier J, Vancanneyt M, Scherer S, Swings J, Cogan TM. Stability of the biodiversity of the surface consortia of Gubbeen, a red-smear cheese. J Dairy Sci. 2007;90:2200–2210. doi: 10.3168/jds.2006-377. [DOI] [PubMed] [Google Scholar]
- Maoz A, Mayr R, Scherer S. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl Environ Microbiol. 2003;69:4012–4018. doi: 10.1128/AEM.69.7.4012-4018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Kodama K, Yasuda H, Okamoto-Kainuma A, Koizumi K, Yamasato K. Presence of halophilic and alkaliphilic lactic acid bacteria in various cheeses. Lett Appl Microbiol. 2007;44:308–313. doi: 10.1111/j.1472-765X.2006.02073.x. [DOI] [PubMed] [Google Scholar]
- Jany JL, Barbier G. Culture-independent methods for identifying microbial communities in cheese. Food Microbiol. 2008;25:839–848. doi: 10.1016/j.fm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Ogier JC, Son O, Gruss A, Tailliez P, Delacroix-Buchet A. Identification of the bacterial microflora in dairy products by temporal temperature-gradient gel electrophoresis. Appl Environ Microbiol. 2002;68:3691–3701. doi: 10.1128/AEM.68.8.3691-3701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Rudolf M, Scherer S. High incidence of Listeria monocytogenes in European red smear cheese. Int J Food Microbiol. 2001;63:91–98. doi: 10.1016/S0168-1605(00)00413-X. [DOI] [PubMed] [Google Scholar]
- Eppert I, Valdés-Stauber N, Götz H, Busse M, Scherer S. Growth reduction of Listeria spp. caused by undefined industrial red smear cheese cultures and bacteriocin-producing Brevibacterium linens as evaluated in situ on soft cheese. Appl Environ Microbiol. 1997;63:4812–4817. doi: 10.1128/aem.63.12.4812-4817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner M, Guenther S, Steffan S, Scherer S. A pediocin-producing Lactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Appl Environ Microbiol. 2003;69:1854–1857. doi: 10.1128/AEM.69.3.1854-1857.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr R, Fricker M, Maoz A, Scherer S. Anti-listerial activity and biodiversity of cheese surface cultures: influence of the ripening temperature regime. Eur Food Res Technol. 2004;218:242–247. doi: 10.1007/s00217-003-0863-4. [DOI] [Google Scholar]
- Ryser ET, Maisnier-Patin S, Gratadoux JJ, Richard J. Isolation and identification of cheese-smear bacteria inhibitory to Listeria spp. Int J Food Microbiol. 1994;21:237–246. doi: 10.1016/0168-1605(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Carnio MC, Eppert I, Scherer S. Analysis of the bacterial surface ripening flora of German and French smeared cheeses with respect to their anti-listerial potential. Int J Food Microbiol. 1999;47:89–97. doi: 10.1016/S0168-1605(99)00016-1. [DOI] [PubMed] [Google Scholar]
- Carnio MC, Höltzel A, Rudolf M, Henle T, Jung G, Scherer S. The macrocyclic peptide antibiotic micrococcin P-1 is secreted by the food-borne bacterium Staphylococcus equorum WS 2733 and inhibits Listeria monocytogenes on soft cheese. Appl Environ Microbiol. 2000;66:2378–2384. doi: 10.1128/AEM.66.6.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saubusse M, Millet L, Delbès C, Callon C, Montel MC. Application of Single Strand Conformation Polymorphism - PCR method for distinguishing cheese bacterial communities that inhibit Listeria monocytogenes. Int J Food Microbiol. 2007;116:126–135. doi: 10.1016/j.ijfoodmicro.2006.12.024. [DOI] [PubMed] [Google Scholar]
- Goerges S, Mounier J, Rea MC, Gelsomino R, Heise V, Beduhn R, Cogan TM, Vancanneyt M, Scherer S. Commercial ripening starter microorganisms inoculated into cheese milk do not successfully establish themselves in the resident microbial ripening consortia of a South German red smear cheese. Appl Environ Microbiol. 2008;74:2210–2217. doi: 10.1128/AEM.01663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan NM, Ward AC, Beresford TP, Fox TP, Goodfellow M, Cogan TM. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl Environ Microbiol. 2002;68:820–830. doi: 10.1128/AEM.68.2.820-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier J, Monnet C, Jacques N, Antoinette A, Irlinger F. Assessment of the microbial diversity at the surface of Livarot cheese using culture-dependent and independent approaches. Int J Food Microbiol. 2009;133(1-2):31–7. doi: 10.1016/j.ijfoodmicro.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Schubert K, Ludwig W, Springer N, Kroppenstedt RM, Accolas JP, Fiedler F. Two coryneform bacteria isolated from the surface of French Gruyere and Beaufort cheeses are new species of the genus Brachybacterium: Brachybacterium alimentarium sp. nov. and Brachybacterium tyrofermentans sp. nov. Int J Syst Bacteriol. 1996;46:81–87. doi: 10.1099/00207713-46-1-81. [DOI] [PubMed] [Google Scholar]
- Callon C, Duthoit F, Delbès C, Ferrand M, Le Frileux Y, De Crémoux R, Montel MC. Stability of microbial communities in goat milk during a lactation year: Molecular approaches. Syst Appl Microbiol. 2007;30:547–560. doi: 10.1016/j.syapm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Irlinger F, Morvan A, El Solh N, Bergere JL. Taxonomic characterization of coagulase-negative staphylococci in ripening flora from traditional French cheeses. Syst Appl Microbiol. 1997;20:319–328. [Google Scholar]
- Bockelmann W, Krusch U, Engel G, Klijn N, Smit G, Heller KJ. The microflora of Tilsit cheese. Part 1. Variability of the smear flora. Nahrung. 1997;41:208–212. doi: 10.1002/food.19970410405. [DOI] [Google Scholar]
- Place RB, Hiestand D, Gallmann HR, Teuber M. Staphylococcus equorum subsp linens, subsp nov., a starter culture component for surface ripened semi-hard cheeses. Syst Appl Microbiol. 2003;26:30–37. doi: 10.1078/072320203322337281. [DOI] [PubMed] [Google Scholar]
- Foulquié Moreno MR, Sarantinopoulos P, Tsakalidou E, De Vuyst L. The role and application of enterococci in food and health. Int J Food Microbiol. 2006;106:1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Franz CM, Stiles ME, Schleifer KH, Holzapfel WH. Enterococci in foods - a conundrum for food safety. Int J Food Microbiol. 2003;88:105–122. doi: 10.1016/S0168-1605(03)00174-0. [DOI] [PubMed] [Google Scholar]
- Collins MD, Hutson RA, Falsen E, Sjödén B. Facklamia tabacinasalis sp. nov., from powdered tobacco. Int J Syst Microbiol. 1999;49:1247–1250. doi: 10.1099/00207713-49-3-1247. [DOI] [PubMed] [Google Scholar]
- Delbès C, Ali-Mandjee L, Montel MC. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl Environ Microbiol. 2007;73:1882–1891. doi: 10.1128/AEM.01716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantsis-Zacharov E, Halpern M. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl Environ Microbiol. 2007;73:7162–7168. doi: 10.1128/AEM.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Ide H, Osaki M, Sekizaki T. Identification of Facklamia sourekii from a lactating cow. J Vet Med Sci. 2006;68:1225–1227. doi: 10.1292/jvms.68.1225. [DOI] [PubMed] [Google Scholar]
- Mounier J, Goerges S, Gelsomino R, Vancanneyt M, Vandemeulebroecke K, Hoste B, Brennan NM, Scherer S, Swings J, Fitzgerald GF, Cogan TM. Sources of the adventitious microflora of a smear-ripened cheese. J Appl Microbiol. 2006;101:668–681. doi: 10.1111/j.1365-2672.2006.02922.x. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Nakajima K, Yanagi M, Yamamoto Y, Yamasato K. Marinilactibacillus psychrotolerans gen. nov., sp nov., a halophilic and alkaliphilic marine lactic acid bacterium isolated from marine organisms in temperate and subtropical areas of Japan. Int J Syst Evol Microbiol. 2003;53:711–720. doi: 10.1099/ijs.0.02446-0. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Tanasupawat S, Nakajima K, Kanamori H, Ishizaki S, Kodama K, Okamoto-Kainuma A, Koizumi Y, Yamamoto Y, Yamasato K. Alkalibacterium thalassium sp. nov., Alkalibacterium pelagium sp. nov., Alkalibacterium putridalgicola sp. nov. and Alkalibacterium kapii sp. nov., slightly halophilic and alkaliphilic marine lactic acid bacteria isolated from marine organisms and salted foods collected in Japan and Thailand. Int J Syst Evol Microbiol. 2009;59:1215–1226. doi: 10.1099/ijs.0.65602-0. [DOI] [PubMed] [Google Scholar]
- Leclercq-Perlat MN, Oumer A, Bergere JL, Spinnler HE, Corrieu G. Growth of Debaryomyces hansenii on a bacterial surface-ripened soft cheese. J Dairy Res. 1999;6:271–281. doi: 10.1017/S0022029999003362. [DOI] [Google Scholar]
- Brennan NM, Cogan TM, Loessner M, Scherer S. In: Cheese: Chemistry, Physics and Microbiology. 3. Fox PF, McSweeney PLH, Cogan TM, Guinee TP, editor. Vol. 2. Amsterdam: Elsevier Academic Press; 2004. Bacterial Surface-ripened Cheeses; pp. 199–225. full_text. [Google Scholar]
- Mounier J, Irlinger F, Leclercq-Perlat MN, Sarthou AS, Spinnler HE, Fitzgerald G, Cogan TM. Growth and colour development of some surface ripening bacteria with Debaryomyces hansenii on aseptic cheese curd. J Dairy Res. 2006;73:441–448. doi: 10.1017/S0022029906001919. [DOI] [PubMed] [Google Scholar]
- Mounier J, Rea MC, O'Connor PM, Fitzgerald GF, Cogan TM. Growth characteristics of Brevibacterium, Corynebacterium, Microbacterium, and Staphylococcus spp. isolated from surface-ripened cheese. Appl Environ Microbiol. 2007;73:7732–7739. doi: 10.1128/AEM.01260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine L, Malcolm GB, Brooks JB, Daneshvar MI. Physiological studies on the growth and utilization of sugars by Listeria species. Can J Microbiol. 1989;35:245–254. doi: 10.1139/m89-037. [DOI] [PubMed] [Google Scholar]
- Premaratne RJ, Lin WJ, Johnson EA. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl Environ Microbiol. 1991;57:3046–3048. doi: 10.1128/aem.57.10.3046-3048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HN, Hodgson DA. Development of a synthetic minimal medium for Listeria monocytogenes. Appl Environ Microbiol. 2003;69:6943–6945. doi: 10.1128/AEM.69.11.6943-6945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lungu B, Ricke SC, Johnson MG. Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions: A review. Anaerobe. 2009;15:7–17. doi: 10.1016/j.anaerobe.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D. Cytoplasmic steps of peptidoglycan biosynthesis. Microbiol Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- Sentandreu R, Northcote DH. Yeast cell-wall synthesis. Biochem J. 1969;115:231–240. doi: 10.1042/bj1150231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain T, Chatterjee SS, Ghai R, Kuenne CT, Billionm A, Steinweg C, Domann E, Kärst U, Jänsch L, Wehland J, Eisenreich W, Bacher A, Joseph B, Schär J, Kreft J, Klumpp J, Loessner MJ, Dorscht J, Neuhaus K, Fuchs TM, Scherer S, Doumith M, Jacquet C, Martin P, Cossart P, Rusniock C, Glaser P, Buchrieser C, Goebel W, Chakraborty T. Pathogenomics of Listeria spp. Int J Med Microbiol. 2007;297:541–557. doi: 10.1016/j.ijmm.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Chavagnat F, Haueter M, Jimeno J, Casey MG. Comparison of partial tuf gene sequences for the identification of lactobacilli. Microbiol Lett. 2002;2:177–183. doi: 10.1111/j.1574-6968.2002.tb11472.x. [DOI] [PubMed] [Google Scholar]
- Kuhnert P, Capaul SE, Nicolet J, Frey J. Phylogenetic positions of Clostridium chauvoei and Clostridium septicum based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1996;4:1174–1176. doi: 10.1099/00207713-46-4-1174. [DOI] [PubMed] [Google Scholar]
- Oberreuter H, Charzinski J, Scherer S. Intraspecific diversity of Brevibacterium linens, Corynebacterium glutamicum and Rhodococcus erythropolis based on partial 16S rDNA sequence analysis and Fourier-transform infrared (FT-IR) spectroscopy. Microbiology. 2002;148:1523–1532. doi: 10.1099/00221287-148-5-1523. [DOI] [PubMed] [Google Scholar]
- Liebgott PP, Joseph M, Fardeau ML, Cayol JL, Falsen E, Chamkh F, Qatibi AA, Labatt M. Clostridiisalibacter paucivorans gen. nov., sp nov., a novel moderately halophilic bacterium isolated from olive mill wastewater. Int J Syst Evol Microbiol. 2008;58:61–67. doi: 10.1099/ijs.0.65182-0. [DOI] [PubMed] [Google Scholar]