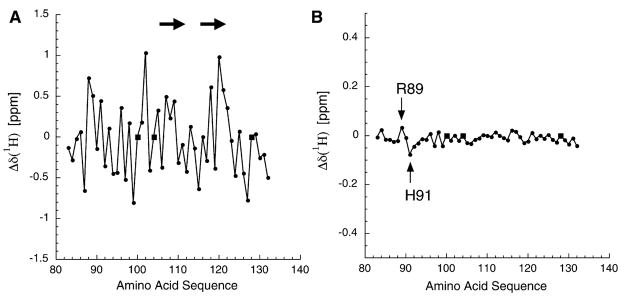
Figure 2. Analysis of AgRP(83–132) 1H Chemical Shifts Derived from Both the Full-Length Protein and the Isolated C-Terminal Domain.
(A) The measured 1H chemical shifts from AgRP(83–132), minus the consensus random coil chemical shifts. The significant variation is consistent with a folded domain. Circles represent measured values; squares represent proline, which is not observable in the HSQC spectra. The arrows show the locations of β strands adjacent to the RFF triplet (residues 111–113).
(B) The difference in chemical shifts, full-length minus C-terminal, for residues 83–132. Note that the vertical axis is expanded by a factor of three compared to that in (A). The limited scatter demonstrates that the HSQC from residues 83–132 in full-length AgRP is essentially equivalent to that of the isolated C-terminal domain, AgRP(83–132). The 2 residues that do show significant 1H chemical shift variations are Arg89 and His91, as indicated.