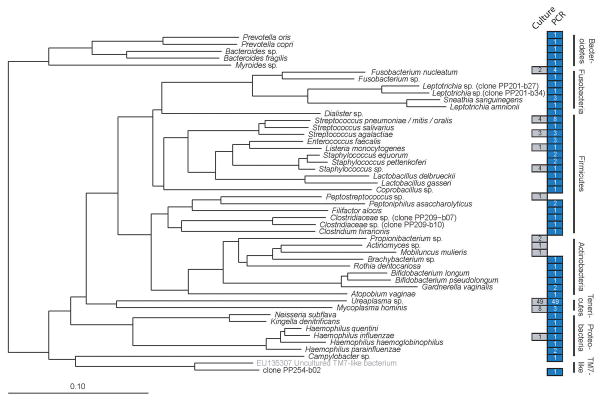
Figure 2. Bacterial taxa detected by PCR or culture.
Phylogeny of the bacterial taxa identified in this study, based on a neighbor-joining algorithm with Felsenstein correction and a 682-column filter. The scale bar represents evolutionary distance (10 substitutions per 100 nucleotides). The taxon in brackets and gray type (uncultured TM7-like bacterium) is a public database sequence included for reference and was not detected in this study. Colored boxes indicate the number of subjects who were positive for a given taxon by culture (gray) or PCR (blue) (some samples were polymicrobial). Because culture isolates were not sequenced, each is represented by a GenBank sequence that corresponds to the taxonomic resolution to which culture isolates were phenotypically identified. Two culture isolates are not represented because they were not characterized to a sufficiently narrow taxonomic resolution to allow tree placement (viridans group streptococcus, and gram positive bacillus, each detected in a separate subject). Candida was the lone fungal genus detected in the study population (not shown in this figure).