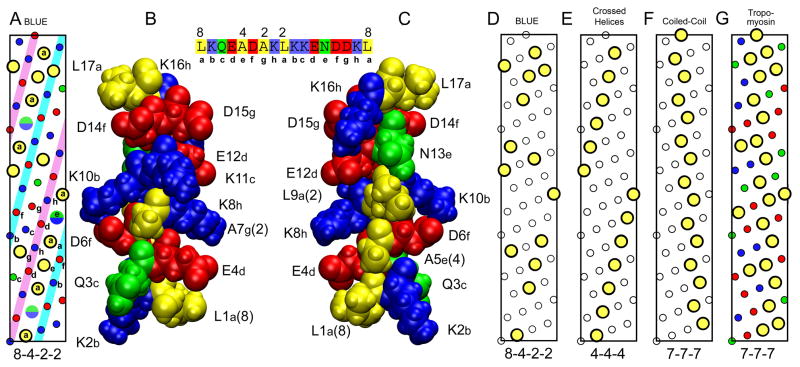
Fig. 6. Helical nature of BLUE-32 and comparison with tropomyosin.
(A) Helical net diagram of hydrophobic residues in BLUE peptide sequence, residues 7036–7088. Large yellow circles are hydrophobic residues with the 8-4-2-2 pattern. The split green/blue circles are the locations of the As or Lys residues between the two leucines separated by 8 residues in the 8-4-2-2 pattern. Small colored circles are charged (red and blue) and polar (green) residues. (B) Space filling model of the BLUE-16 repeat. Structure of BLUE16 was generated by ROSETTA and then subjected to 4 ns of MD in a water box at low ionic strength. Structure orientated to show the residues QAKD that are in the white strip on the left side of the inset helical net between the two structures. (C) Same BLUE-16 structure rotated 180 degrees to show the hydrophobic strip as in the helical net, residues LALNL. Sequence of peptide with 8-4-2-2 repeat pattern shown at top between structures. (D) The 8-4-2-2 pattern is shown with only the location of the hydrophobic residues highlighted for clarity. (E) Helical net typically found in globular proteins.25 (F) Helical net found in coiled-coil proteins such as tropomyosin. (G) Helical net for residues 61–113 of porcine tropomyosin that the BLUE-32 peptide threaded to.