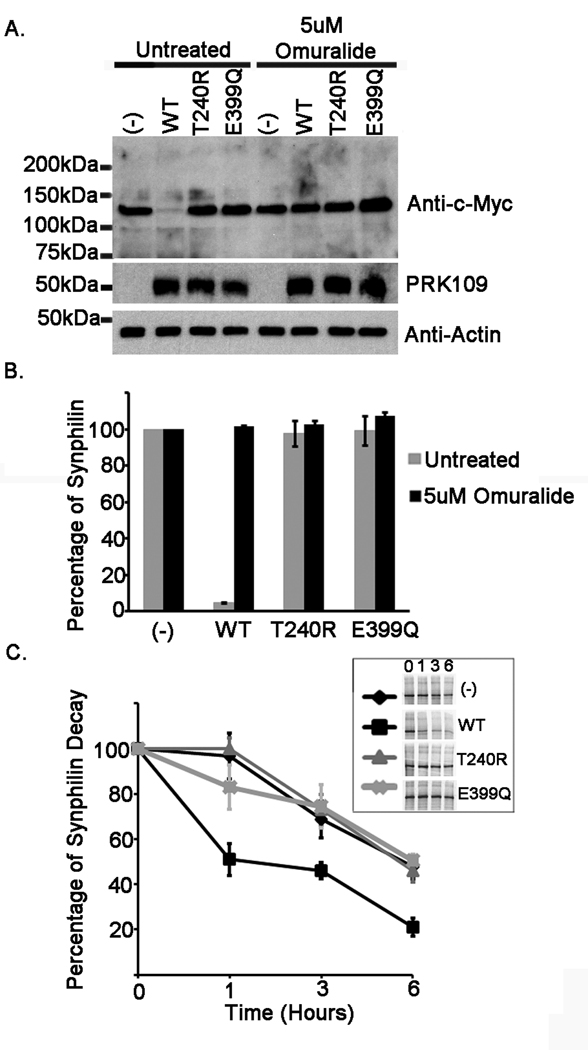
Figure 7.
E399Q mutant parkin is functionally impaired. HEK293T cells were co-transfected either with mock pcDNA3.1 vector “(−)” or full length WT, T240R, or E399Q human parkin pcDNA 3.1 constructs as well as the pcDNA3.1 myc-tagged full-length synphilin-1 construct. The co-transfection experiments were performed using a parkin to synphilin-1 cDNA ratio of 4:1. At 24 hours post transfection, (A and B) cells were incubated for 16 hours with fresh DMEM or with DMEM containing 5uM Omuralide. Cells were then harvested and total protein lysates were extracted. Extracts were resolved by 10 % polyacrylamide gels and immunoblotted with the monoclonal antibodies anti-c-Myc clone 9E10 to assess synphilin-1 levels, PRK109 antibody to confirm parkin expression, and anti-actin antibody to ensure equal protein loading. All experiments were performed in triplicate and repeated at least 3 times. The mobility of molecular mass markers is indicated on the left. The levels of synphilin-1 were quantified as described in “Materials and Methods” and the graph in B depicts the percentage of synphilin-1 protein standardized to that in the mock vector samples. The error bars indicate standard deviation between replicate samples (n=4). C) Cells were pulsed with 35S-methionine for 1 hour and chased for 0, 1, 3, or 6 hours. The inset shows representative pulse-chase experiments. Experiments were conducted in triplicates. The results are plotted as percentage of protein over time standardized to the 0 hrs time point. The error bars show standard deviation (n=3).