Abstract
Intracellular signaling cascades are a series of regulated protein-protein interactions that may provide a number of targets for potential drug discovery. Here, we examine the interaction of Regulators of G protein signaling (RGS) proteins with the G protein Gαo, using a flow cytometry protein interaction assay (FCPIA). FCPIA accurately measures nanomolar binding constants of this protein-protein interaction, and has been used in high throughput screening. This report focuses on five RGS proteins (4, 6, 7, 8 and 16). In order to increase the content of screens, we assessed high throughput screening of these RGS proteins in multiplex, by establishing binding constants of each RGS with Gαo in isolation, and then in a multiplex format with five RGS proteins present. In order to use this methodology as a higher-content multiplex protein-protein interaction screen, we established Z' factor values for RGS proteins in multiplex of 0.73 to 0.92, indicating this method is suitable for screening using FCPIA. To increase throughput, we also compressed a set of 8,000 compounds by combining 4 compounds in a single assay well. Subsequent deconvolution of the compounds mixtures verified the identification of active compounds at specific RGS targets in our mixtures using the polyplexed FCPIA method.
Keywords: G protein, RGS, Flow Cytometry, FCPIA, High throughput screening
INTRODUCTION
Inhibition of protein-protein interactions represents a unique approach to disrupting intracellular signaling networks. Technically, directly measuring protein-protein interactions has been difficult, often relying on fluorescence polarization (FP) assays, which use a representative peptide and one complete protein moiety to assess the interaction. Other often used methods are surface Plasmon resonance (SPR), TR-FRET and Alphascreen which also have significant limitations with regard to protein size and purity, as well as a number of intrinsic experimental factors, such as buffer composition, that can effect the signal quality. The strength of signal may also be of issue in both FP and SPR methods, which does not necessarily render them uninformative, but significant optimization of protocols is often necessary. Furthermore, some limitations for these methods exist when considering the goal of discovering new small molecule inhibitors of specific protein-protein interactions, in that they can require significant amounts of protein and peptide material in the case of FP, or may be far too slow for serious consideration for high throughput screening, in the case of SPR. In addition, TR-FRET and Alphascreen require reagents, such as terbium or europium chelates that add significantly to assay cost. Successful cell-based assays have been developed for screening for inhibitors protein-protein interactions, using luciferase as a reporter, but an isolated protein-protein interaction can be difficult to distinguish in these systems, and off-target effects may be present (2, 3). Due to the aforementioned limitations of existing methodologies, our group and others have had longstanding interest in the implementation of flow cytometry for investigating protein-protein and protein-ligand interactions (1, 4–9). Recent publication of a flow cytometry protein-protein interaction assay (FCPIA) developed as a high throughput screening method for screening for inhibitors of Regulator of G protein signaling 4 (RGS4) is exemplar of the feasibility of using FCPIA to mine for RGS/Gαo interaction inhibitors (1).
Regulators of G protein signaling (RGS) proteins are intracellular modulators of G protein-coupled receptor signaling (10) and represent intriguing drug targets (11,12). While cell-surface expressed ligand-binding domains, such as receptors and ion channels are commonly exploited targets, the inhibition of signal at the initiation of cascades usually results in complete shutdown of the signal. In contrast, inhibition of downstream protein-protein interactions allows manipulation of discrete steps of a signaling cascade that may result in more subtle and specific effects (13–15).
We are interested in chemically targeting the key regulatory interaction between RGS proteins and Gα subunits. Classically, the signals mediated by G protein coupled receptors (GPCRs) upon ligand binding are mediated through heterotrimeric G proteins. Upon ligand binding and receptor activation, G protein α and αγ subunits dissociate from receptor and modulate the activity of a number of downstream effectors, ranging from ion channels to adenylate cyclase (16). The activation of the G protein is dependent on the α subunit, which exists in a GDP liganded form in its inactive state and undergoes rapid GTP for GDP exchange upon activation. Inactivation of these G protein signaling pathways occur when the bound GTP is hydrolyzed back to GDP and the Gα subunit returns to its inactive, GDP-liganded form. Gα subunits possess intrinsic GTPase activity but this process is too slow to account for physiological processes such cardiac and visual signal transduction (17, 18). Physiological rates of G protein inactivation are achieved through the action of RGS proteins (19, 20). RGS proteins act as GTPase Accelerating Proteins (GAPs) which shorten the lifetime of the active Gα and Gβγ proteins from minutes to milliseconds or seconds (21).
A large percentage of therapeutic drugs target GPCRs, but we are interested in examining the RGS/Gα interaction as a unique target, because inhibiting an RGS could have several interesting physiological effects and perhaps better selectivity ((for review, see 12, 22, 23)). For instance, an RGS inhibitor could potentiate the action of natural or exogenous agonist ligands. Specificity could also be introduced, relying on the discrete tissue expression patterns of some RGS proteins. For these reasons, our laboratory and others have been interested in modulating the RGS/Gα interaction, with successes ranging from our cyclic octapeptide and small molecule RGS inhibitors, to a small molecule that stabilizes an RGS/G protein interaction, as well inhibitors identified through a yeast two-hybrid screen (1, 24–27). Our current goal is to expand on the high throughput capabilities of the FCPIA method, by increasing the content of experiments and future screens to examine multiple RGS/G protein interactions simultaneously.
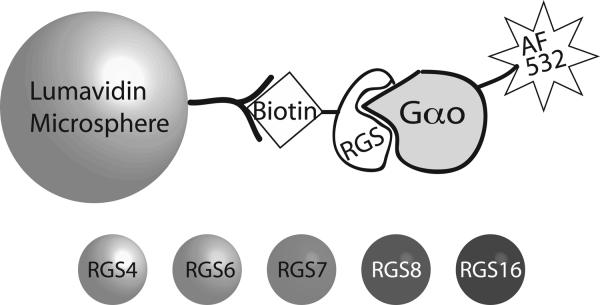
FCPIA consists of three key components (Figure 1) 1) avidin-coated microspheres, 2) biotinylated RGS, and 3) activated Gαo labeled with AlexaFluor 532. Experimentally, biotinylated RGS coupled to avidin-coated beads is co-incubated in a 96 well plate with activated Gαo. As samples from each well are aspirated into the Luminex flow cytometer, the bead is detected and bead-associated fluorescence is measured, providing a quantitative reading of the amount of Gαo bound to RGS (1). In this study, we advance our ability to make quantitative protein-protein interaction measurements in a multiplexed format. Lumavidin beads are available with 100 different internal dye-ratios (regions), which the Luminex flow cytometer can distinguish in the flow cell. Therefore, multiple RGS proteins can be coupled to individual bead regions (Figure 1), and then mixed together to perform binding analyses to a common, fluorescently labeled binding partner, in this case Gαo. In this study, we compare affinity measurements of 5 different RGS proteins to Gαo in individual (singleplex) to multiplex measurements. In addition, we established Z-factor values for a five-plex RGS system to use in a high throughput FCPIA paradigm, demonstrating the suitability of multiplexing protein-protein interaction measurements to measure individual protein-protein interaction affinities as well as for use in higher-content FCPIA.
Figure 1.
Diagram of the components of FCPIA. Avidin-coated microspheres are incubated with biotinylated RGS protein to yield RGS-coated beads. 5 different bead regions are utilized, with each RGS being bound to a unique bead region. These bead regions are discriminated in the Luminex analyzer, and associated fluorescently labeled Gαo is detected. An inhibitor would result in the disruption of the RGS/Gαo interaction, and detected as a loss of bead-associated fluorescence.
In this study, we introduce a new paradigm to screening small molecule libraries using flow cytometry. Due to the limitations of technology, in terms of throughput speed, we report the utilization of what we term a “polyplexed screen.” In this paradigm, we have used 5 different RGS proteins on 5 different Luminex bead regions, allowing us to multiplex our 5 RGS targets in a single well. We chose these 5 RGS proteins because they belong to two separate RGS protein families (R4 and R7) to allow for the identification of RGS-family inhibitors, as well as being heterologously expressed in good quantity from E. coli. For the RGS7 family members RGS7 and RGS6, we expressed the RGS Homology Doman (RH), as it is the minimal unit necessary to interact with Gαo and is stably expressed in E. coli. In order to increase the number of compounds that could be screened, we also performed a 4:1 compression of an 8,000 compound subset of the Maybridge HitFinder library held by the University of Michigan Center for Chemical Genomics (CCG). For this compression, 4 compounds were deposited into each test well of a 96-well plate (80 wells, 16 used for controls), resulting in 320 compound entities per screening plate. Upon screening, 5 targets on beads were added to each well, increasing our data density to 20 data points per well (5 targets by 4 compounds), or 1600 data points per 96-well plate, a marked increase in the content of our screen from the normal 80 data points.
Due to the design of this prototype screen, it was necessary to perform a deconvolution of any hit wells in order to test our ability to detect and identify one active compound within a four compound mixture. After our initial screen and dose-response follow-up, we were able to identify active individual compounds from the 4 compound mixtures, the majority of which exhibited one active compound per well, with only 2 exceptions.
In this study, we explore the feasibility of performing 5 target multiplexing using the flow cytometry protein interaction assay (FCPIA) as well as using 4 compound mixtures as our screening library. We demonstrate the ability of this method to increase content with little time penalty and the robustness of the method in identifying a single active compound from our screening mixtures (Table 2).
Table 2.
Multiplex Screen Summary, 8,000 Compounds
| Active Wells | Unique Wells | Cmpds with Activity in DRC | Number of unique wells with confirmed actives | |
|---|---|---|---|---|
| RGS 4 | 44 (2.2%) | 15 (0.75%) | 9/60 (15%) | 7 |
| RGS 8 | 65 (3.3%) | 7 (0.35%) | 2/28 (7%) | 2 |
| RGS 16 | 77 (3.9%) | 27 (1.4%) | 14/104 (13%) | 10 |
| RGS 6 | 29 (1.5%) | 19 (0.95%) | 3/76 (4%) | 3 |
| RGS 7 | 34 (1.7%) | 6 (0.3%) | 2/24 (8%) | 2 |
In the table above, the number (percentage) of active wells was defined as those wells showing a 60% or greater inhibition of the RGS/Gαo interaction for the indicated RGS. The number of unique wells indicates the number of wells for which only the single specified RGS was inhibited. Those unique wells were further analyzed in DRC and the number compounds with activity following deconvolution of the four compounds from each active well is shown. The number of unique wells that produced an active compound is indicated in the last column. Thus for RGS4 and RGS16 some wells produced more than one active compound but for RGS6, 7, and 8 there was only one active compound (out of the four) in each well.
METHODS
Protein Expression, Purification and Labeling
RGS16
human RGS16 in pcDNA3.1(+) vector was purchased from the UMR cDNA resource (www.cdna.org). The open reading frame containing full-length RGS16 was amplified using the primers: 5'-GAATTCATGTGCCGCACCCTGGCCGC-3' and 5'-GTCGACGGTGTGTGAGGGCTCGTCCA-3', which amplify the entire open reading frame and add compatible EcoNI and SalI restriction enzyme recognition sites on the 5' and 3' ends, respectively. The fragment was amplified using standard PCR conditions, gel-purified as per manufacturer's protocol (Qiagen, Valencia CA) and ligated into Antarctic phosphatase treated pMALC2H10T vector digested with EcoNI and SalI. The ligated product was transformed into DH5α bacteria, colonies picked, miniprepped (Qiagen, Valencia CA) and sequenced to verify in-frame placement of RGS16 to form the maltose binding protein (MBP)-RGS fusion protein.
RGS6
The human RGS6 RGS homology domain (RH, box) was cloned from total human brain mRNA (Clontech, Mountain View CA) using reverse transcriptase polymerase chain reaction (RT-PCR). The primers used for RT-PCR were designed to add restriction sites, BamHI and SalI for cloning into pMALC2H10T were 5'-ATCCGAAAATGTCGACGTTTGAAGA-3' and 5'-AAGAAGAGAGGGCTGTAAACAGGAG-3' RT-PCR was performed using SuperScript™ One-Step RT-PCR with Platinum® Taq from Invitrogen (Carlsbad, CA) as per the manufacturer's protocol using a 55C annealing temperature. The RT-PCR resulted in a ~600bp product, detected by agarose gel electrophoresis and purified using a gel-purification kit (Qiagen, Valencia CA). The PCR product was ligated into Antarctic phosphatase treated pMALC2H10T expression vector cut with BamHI and SalI. The sequence was verified to be identical with the RGS6 box by sequence alignment with sequence available in the NCBI database.
RGS7 and RGS8
RGS7 RH domain, and full-length RGS8 were purified as GST-fusion proteins as previously described (4).
RGS4
An N-terminally truncated (ΔN51) RGS4 pMALC2H10T DNA construct was a kind gift from Dr. John Tesmer (University of Michigan). This construct expresses the RGS4 as a C-terminal fusion on the bacterial MBP. The construct expresses at much higher levels than other HIS-tagged or native RGS4 constructs however, it binds Gαo with slightly lower affinity than a previously published ΔN18-RGS4 construct (1).
Generation of Cysless RGS4 Mutant
The following oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and used in the Quick-Change Multi Site-Directed Mutagenesis Kit according to the manufacturer's protocol (Stratagene, La Jolla CA). Mutagenesis was required to be completed in two rounds, due to C197 and C204 being localized too close to each other and causing primer overlap. The primer sequences are: C71A 5'-GCTGGAAAACCTGATTAACCATGAAGCTGGACTGGCAGCT-3', C95A 5'-GAACATTGACTTCTGGATCAGCGCTGAGGAGTACAAGAAAATCAAA-3', C132A 5'-GAGGTGAACCTGGATTCTGCCACCAGAGAGGAGACAAG-3', C148A 5'-GTTAGAGCCCACGATAACCGCTTTTGATGAAGCCCAGAAG-3', C183A 5'-CCAATCCTTCCAGCGCCGGGGCAGAGAAGC-3', C197A 5'-CCAAGAGTTCTGCAGACGCCACTTCCCTAGTCCCTC-3', C204A 5'-TTCCCTAGTCCCTCAGGCTGCCTAATTCTCACACA-3'.
Gαo
Rat Gαo was purified as an N-terminal 6X-HIS tagged construct expressed in E. coli as previously described (28). Gαo activity was assessed using GTPγ35S binding (29).
General MBP-RGS Fusion Protein Purification
For bacterial expression of MBP fusion proteins, vector was transformed into BL21(DE3) bacteria and a single colony was placed in 100mL of Enriched Media with glucose (EMG) overnight, shaking at 37C. The next morning, 6L of EMG was inoculated with the starter culture, and grown to OD600=0.6. 100μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added and induction proceeded for 4h at 37C. Bacteria were then pelleted, flash-frozen in liquid nitrogen and stored at −80C until purification. Briefly, frozen pellets were thawed in 50mM HEPES, 100mM NaCl and 1mM DTT pH 7.2 (MBP buffer) with added protease inhibitors (E-64, phenylmethanesulfonylfluoride (PMSF), lima bean trypsin inhibitor, TPCK) and treated with 0.8mg/mL lysozyme for 20min. Following lysis, 1mM (final) MgCl2 and 8ug/mL DNAse I were added while stirring. After 10min, the lysate was centrifuged for 40 min at 35,000 RPM (~110,000 × g) in a Ti40 (Beckman) rotor. The supernatant was filtered (0.45μm) and applied to a 10mL amylose column (New England Biologicals). The column was washed with 200mL MBP buffer and 1mL fractions were eluted with 10mM maltose in MBP buffer. Approximately 80% pure intact fusion protein eluted in fractions 3–12. Fractions were pooled, concentrated using an Amicon Ultra concentrator (Millipore) and snap frozen in liquid nitrogen.
Chemical Biotinylation of Purified RGS Proteins
RGS proteins were biotinylated with amine-reactive N-(+)-Biotinyl-6-aminocaproic acid N-succinimidyl ester (Biotin-NHS, Fluka 14412) in a 3:1 (biotin:RGS) stoichiometry as previously described (1).
Flow Cytometry Protein Interaction Assay (FCPIA)
RGS Single and Multiplex Saturation Assay
Experiments were carried out in 96-well conical-bottom PCR plates and samples were read using a Luminex 200 bead analyzer. The protocol used for single-RGS saturation experiments has been previously described (1). Briefly, Luminex LumAvidin beads (500 per RGS subtype per well) were vortexed, briefly sonicated and diluted into 1mL Bead Coupling Buffer (BCB) (PBS, pH 8.0 supplemented with 1% BSA). The beads were pelleted (60s at 7K RPM) supernatant removed, and resuspended in 1mL BCB. This process was repeated for 3 washes. The beads were then resuspended in 500μL BCB and biotinylated RGS protein added at 20× the final desired concentration (10nM, final). The beads were incubated for 30 min at RT. After the bead coupling was complete, the beads were spun down, washed with 1mL BCB 3 times. For multiplex assays, at this point the RGS-beads were combined and resuspended in 5mL of Flow Buffer (50mM HEPES, 100mM NaCl, 0.1% Lubrol, 1% BSA pH 8.0). 50μl of beads were dispensed into each well of the 96 well plate and incubated at RT for 10 min. Blank beads (i.e. bead coupling reaction conditions in the absence of RGS) were used as controls in some experiments to determine nonspecific binding. During the 10 minute incubation of RGS with compound, the AF532-labeled Gαo solution was prepared. GDP (5μL 10mM), MgCl2 (500μl, 50mM), AlCl3 (500μL, 50μM), and NaF (500μL, 50mM) were added to 3.5mL flow buffer and AF532-Gαo was added to form the activated Gαo-GDP-AlF4- complex (at 200nM final concentration), which binds RGS with high affinity. This solution was then diluted serially down the 96 well plate to yield final Gαo concentrations between 0.78 and 100nM. (Note: for some experiments, the GDP control is Gαo added without the addition of MgCl2, AlCl3 and NaF to yield GDP-bound, and hence very low affinity (background) levels of RGS/Gαo binding.) After 10 min to ensure activation of the Gαo, 50μL was aliquoted to each RGS-bead and compound-containing well of the 96 well screening plate. The proteins were incubated for 30 min at RT in the dark before being read on a Luminex 200IS 96-well plate reading flow cytometer. Median fluorescence intensity values for each bead set (RGS) were calculated and used for data analysis.
Z-factor Determination
Z-factor was determined using the following equation: where σ represents the standard deviation of positive and negative (p, n) controls, and μ represents the mean of positive and negative control values. Positive controls were determined using 48 wells of a 96 well plate, containing 5 RGS proteins on different bead regions at a final concentration of 10nM, and the addition of AMF-treated Gαo (50nM, final). Negative controls are treated in the same fashion, but substituting mock-coupled (“blank”) beads for the RGS-coupled beads.
DNA Sequencing
All DNA constructs were verified by sequencing through the University of Michigan DNA Sequencing Core Facility.
Polyplexed High Throughput Screen
8,000 compounds from the Maybridge HitFinder collection were screened. Briefly, 0.5uL from each of four compound master plates (1.5mM compound concentration in DMSO) were transferred via a Beckman BioMek XL liquid handling robot into the corresponding well of a 96 well assay plate. This resulted in a total of 2uL compound in each well of a 96 well plate. These concentrations kept DMSO concentrations below 4%, a conservative upper limit tolerance for the assay (data not shown). Well registry was preserved, with well A2 from master plates 1–4 being dispensed into well A2 of the assay plate, resulting in 96 well assay plates with four compounds per assay well. The assay format reserved rows 1 and 12 for positive and negative controls, leaving 80 experimental wells per plate that contained the compound mixtures.
Lumavidin microspheres were prepared as described above, and coupled to each of the 5 different RGS proteins. RGS-beads were added to compound-containing plates as a single 50uL aliquot per well. A 10 min incubation at RT was followed by addition of 100nM AlexaFluor 532-labelled Gαo in a 50uL aliquot to yield a 50nM final Gαo concentration. The mixture was incubated in the dark for 30 min at RT, and then read on a Luminex 200 bead analyzer, collecting 75 events per bead region per well. The final screening conditions per 100uL volume in each well were: 50nM AF532-Labeled Gαo, 7.5uM each screening library compound, 2% DMSO, and 10nM each RGS on 500 beads. Positive controls (loss of Gαo binding signal) consisted of wells without MgCl2, AlCl3 and NaF (-AMF), while negative control wells (preserved Gαo binding signal) consisted of the screening mixture without any screening library compound.
Materials
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO), Fisher Scientific (Hampton, NH) or Acros Organics (Geel, Belgium) and were reagent grade or better. Avidin-coated microspheres for flow cytometry were from Luminex (Austin, TX). Molecular biology kits were purchased from sources indicated in the text. Data were analyzed using Graphpad Prism 5.0 (Graphpad Software, San Diego CA).
Fluorescent Labeling of Gαo
Purified Gαo was chemically labeled with AlexaFluor® 532 carboxylic acid succinimidyl ester (Invitrogen, Carlsbad CA) at a 3:1 fluorophore: protein ratio. Purified Gα0 (500ug, 12.5nmol) was diluted in 250μL H50E1N100 (50mM HEPES, 1mM EDTA, 100mM NaCl, pH8) buffer supplemented with 10μM GDP. 2.8μL (~38nmol) of an AF532 solution (1mg/100μL DMSO) was added, and the solution was incubated at 4C in the dark for 1.5 hours. The reaction was quenched with 20μL of 1M glycine and for 30 min. Excess fluorophore was removed via 1mL Sephadex G25 spin column and elution with HEN buffer supplemented with GDP. Activity and effective concentration of Gαo was determined post-labeling using GTPγ35S binding (29).
Flow Cytometry Dose-Response Experiments
Experiments were done similarly to the flow cytometry screening assay, except that the total assay volume was 150μL, with 50μL of RGS4-beads (coupled at 3× final concentration, 6nM), 50μL Gαo-AF532 and varying concentrations of CCG compounds to give a final range of 100 to .01 μM. Final assay concentrations for RGS and Gαo were 2nM and 15nM, respectively.
RESULTS
Effect of Multiplexing RGS Targets
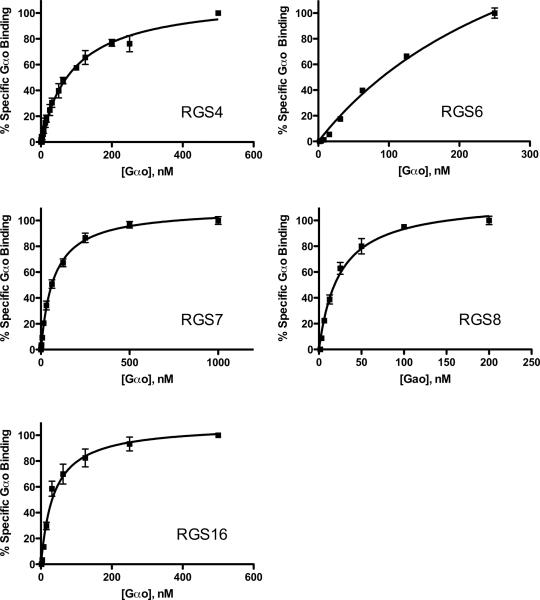
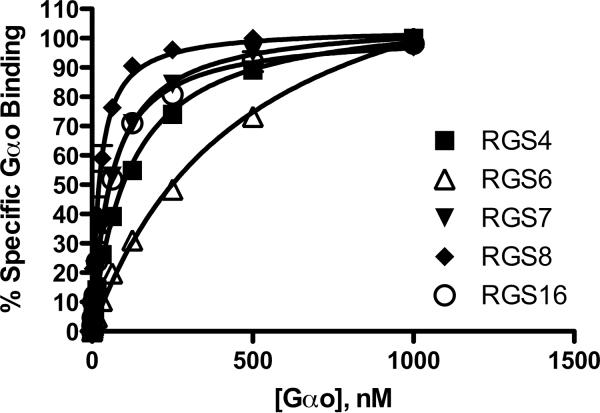
We tested binding affinities between the RGS/Gα pairs in order to establish the multiplexed screening method. We compared the affinity measurements for 5 RGSs with their binding partner, Gαo in both singleplex (one RGS per sample) and multiplex (5 RGS per sample) assays. Nonspecific binding was determined using microspheres without bound RGS, and was subtracted from total binding data to yield specific binding values. Non-specific binding was less than 10% of total, except in the case of RGS6, in which non-specific binding was approximately 15%. Figure 2 depicts the saturation isotherms for individual of measurements of specific binding of the RGS proteins, whereas Figure 3 shows the same measurements made in multiplex. Table 1 summarizes the binding data. Notably, RGS 4, 7, 8 and 16 have Kd values < 100 nM for Gαo, while RGS6 has a 3- to 4-fold lower affinity. Individual Kd values for an individual RGS protein were not significantly different when measured in single and multiplex formats (p >0.05).
Figure 2.
Measurement of RGS-Gαo protein-protein interaction individually. Each panel shows the specific Gαo bound to a particular RGS-bead. Increasing concentrations of fluorescently labeled Gαo were added to RGS on beads (10nM final RGS concentration) in the presence of AlF4− to determine total binding, and in the absence of AlF4− to determine the non-specific component. Non-specific binding was subtracted from total to yield the % Specific Gαo bound to the RGS, with 100% indicating the maximum fluorescence of saturated, Gαo-bound RGS in the presence of GDP-AlF4− as compared to GDP alone). Typical non-specific binding was less than 10% total signal for each RGS, except for RGS6, which had a non-specific signal of up to 15% at the maximal Gαo concentration in saturation experiments. Each graph is the average of at least 3 independent experiments (N=3), performed in duplicate, with error bars indicating the standard error of the mean (SEM).
Figure 3.
Multiplexed measurement of RGS-Gαo protein-protein interaction. Increasing concentrations of fluorescently labeled Gαo were added to 10nM RGS on beads in the presence of AlF4− to determine total binding, and in the absence of AlF4− to determine the non-specific component. In these experiments, each RGS was coupled to an individual bead region, washed, and then the beads mixed together before being added to the test wells. Non-specific binding was subtracted from total to yield the % Specific Gαo bound to the RGS, (% Gαo bound in the presence of GDP-AlF4−as compared to GDP alone). In addition, non-specific binding was tested to each bead region to ensure no intrinsic differences existed. Typical non-specific binding was less than 10% total signal, except for RGS6, which had higher non-specific binding (approx. 15%) at saturating Gαo concentration. Each line on the graph is the average of at least 3 independent experiments (N=3), performed in duplicate, with error bars indicating the standard error of the mean (SEM).
Table 1.
Single and multiplex affinity measurements (Kd ± SEM), nM
| Single | Multiplex | p value | |
|---|---|---|---|
| RGS 4a | 91 ± 9 | 115 ± 6 | 0.06 |
| RGS 8 | 24 ± 2 | 23 ± 2 | 0.7 |
| RGS 16 | 37 ± 6 | 54 ± 7 | 0.1 |
| RGS 6 | 340 ± 52 | 478 ± 27 | 0.08 |
| RGS 7 | 72 ± 5 | 63 ± 2 | 0.2 |
N=5
others N=3
Z-factor Determination
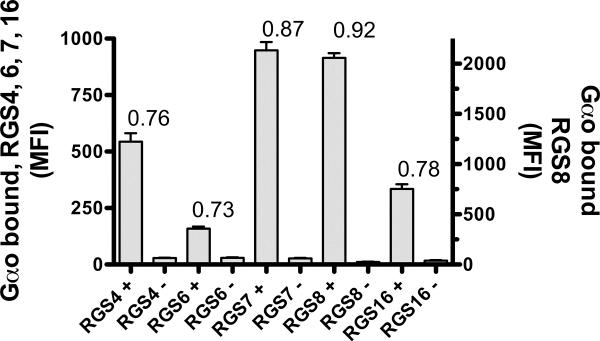
In order to assess the suitability of using multiplexed targets in the FCPIA screen, we determined Z-factors for each RGS-Gαo pair in a multiplexed format. This used a 96-well plate with 5 distinct RGS labeled beads plus soluble AlexaFluor 532-labeled Gαo. Forty-eight wells were used for the positive controls in the presence of AlF4− to stimulate high-affinity RGS-Gαo binding and 48 wells for negative controls in the absence of AlF4− (Figure 4). Z-factors were as follows: 0.76 (RGS4), 0.73 (RGS6), 0.87 (RGS7), 0.92 (RGS8), 0.78 (RGS16). The Z-factors were all well above the value of 0.5 that indicates an assay that is acceptable for high throughput screening (30).
Figure 4.
Z-factor determination. On a 96-well assay plate, 48 wells were designated positive controls, and 48 wells were designated as negative controls. Positive control wells contained AlF4− to induce RGS/Gαo association whereas negative control wells did not contain AlF4−. All wells contained the mixture of 5 RGS proteins (10nM, final) on individual beads (500 beads per RGS) and AlexaFluor 532-labeled Gαo (50nM, final). Average values ± Standard Deviation were plotted, demonstrating the signal strength and consistency (standard deviation) of the controls. Z-factors were calculated using the equation given in the methods section, individual Z-factors are shown above the bars above. Positive and negative control means and standard deviation values used for calculating Z-factors were as follows, 543+37 / 29±2(RGS4) 159±9 / 30±2.5 (RGS6), 948±36 / 27±2 (RGS7), 2057±47/ 26±2 (RGS8), 335±21 / 17±2 (RGS16).
Multiplex Screening Results
Table 2 summarizes the screening results from an 8,000 compound subset of the Maybridge HitFinder collection. The cutoff for defining an active was set at 60% inhibition of the RGS-Gαo interaction as measured using FCPIA. Table 2 depicts the number of active wells for each RGS, as well as the number of unique wells, indicating that a well showed activity for only a single RGS at the 60% cutoff. Subsequent cherrypicking and dose-response experiments were done on each of the 4 compounds from each unique active well, with the indicated number of compounds demonstrating inhibition in the dose-response study. As expected, most wells produced only one active compound from each mixture of 4. Of the 74 unique 4-plex wells examined, 24 produced active compounds (32% confirmation rate) For the 24 wells with activity in the primary screen for which confirmed active compounds were found, 18 wells yielded a single compound while 6 wells yielded two actives for a total of 30 confirmed actives. Thus the compound deconvolution was reasonably successful.
Compounds with Multiple Activities
During dose-response follow up experiments with freshly ordered powders, compounds which were active on RGS4 family members (RGS 4,8 and 16) were tested at all three of the individual RGS members to test for within-family activity. Table 3 summarizes the % inhibition by the compounds on other RGS4 family members at 10μM.
Table 3.
Compounds with Activity on Multiple Targets, as % Inhibition at 10uM
| Compound | RGS 4 | RGS 8 | RGS 16 |
|---|---|---|---|
| CCG-420854 | 33% | 3% | 8% |
| CCG-4388616 | 31% | 3% | 3% |
| CCG-462324a | 3% | 1% | 10% |
| CCG-500144 | 93% | 52% | 90% |
| CCG-510964a | 4% | 3% | 5% |
| CCG-5183116 | 15% | 10% | 15% |
| CCG-540848 | 76% | 9% | 42% |
| CCG-5591916 | 75% | 8% | 18% |
Initial RGS “active”
Initial RGS “active”
Initial RGS “active”
Lower potency at DRC
The table above shows individual compounds that had activity on more than one RGS as discovered in follow-up dose-response experiments. Superscripts after the compounds number indicate the RGS at which the compound was originally identified as having activity, and the percentages indicate the amount of inhibition at 10uM concentration of compound. For two compoundsa, the level of inhibition was quite low at 10uM, but these compounds did show dose-response activity, albeit with considerably higher IC50 values.
Potential Mechanism of RGS Inhibitors
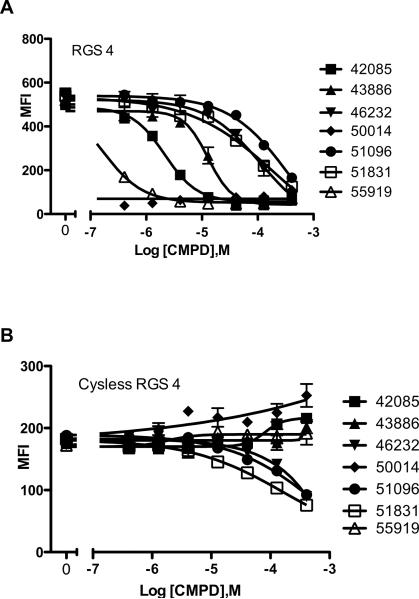
In order to assess the potential role of cysteine modification in the mechanism of action of the 7 identified RGS4 inhibitors, we tested the compounds at an RGS4 construct with each of its 7 cysteine residues mutated to alanine. Figure 5 shows the loss of potency for inhibition of RGS4 by each of the 7 compounds at the cysless RGS4 mutant, indicating the likelihood that these compounds may act to alter the activity of RGS through the covalent modification of cysteine residues.
Figure 5.
RGS4 hit compound dose-response curves and mechanistic evaluation. Panel A shows the effect of increasing compound concentrations on the ability of RGS4 (2nM on 500 beads, final) to associate with Gαo (15nM, final), as reflected by a loss of the Median Fluorescence Intensity (MFI) or bead-associated fluorescence. Panel B shows the same compounds' effect on an RGS4 mutant with all 7 cysteines mutated to alanine. A marked loss of potency for each of the compounds indicates a mechanism potentially involving the modification of cysteines in RGS4, which can inhibit RGS4 function.
DISCUSSION
The potential utility of small molecules that disrupt the function of specific RGS proteins is manifold. Mechanistically, such inhibitors could be useful to increase the potency and selectivity of current GPCR agonist therapeutics, and may provide interesting pharmacological effects on their own (12, 22, 23, 31).
The current state of discovery efforts for RGS inhibitors has yet to identify a specific, potent, cell-permeant and functional small molecule RGS inhibitor, but progress has been made in identifying ligands that can bind and disrupt RGS function in several systems. Initially, our lab identified a cyclic octapeptide based on the Switch 1 region of the RGS/Gαil crystal structure that inhibits the GAP activity of RGS4 (24, 25, 32). Another group identified compounds in a yeast-based screen, but no structural information or further report has been available (26). Another small molecule, CCG-4986 was identified in our laboratory, appears to inhibit RGS4 function through irreversible cysteine modification, and does not inhibit RGS4 in intact cell systems, likely due to the reducing glutathione present in cells (1).
This study aimed to increase our ability to rapidly screen small molecule libraries for compounds that inhibit RGS function by disrupting its interaction with the G protein Gαo. FCPIA provides a platform that allows us to multiplex our targets, as with the 5 RGS proteins screened in the assays described here. First, we demonstrated that the accurate measurement of these protein-protein interaction pairs could be accomplished in multiplex (Figures 2&3, summarized in Table 1). These saturation experiments show that the presence of other RGS proteins in the mixture does not interfere with our ability to measure specific interaction. The saturation experiments also allowed us to assess the relative Kd values for each RGS/Gαo interaction. We chose a screening concentration of Gαo (50nM) that was in the linear portion of the RGS/Gαo interaction saturation curves. By choosing this concentration, we increased the likelihood of detecting a disruption of the RGS/Gαo interaction.
These screens took place at a time when the limit of our available technology only allowed us to screen approximately two 96-well plates per hour, and we sought to increase the content value of our screens by multiplexing targets and compressing a small library to four compounds per well, resulting in 20 data points per experimental well. This increase allowed for the relatively slow (by HTS standards) FCPIA to be amenable to screening.
The results of our 8,000 compound screens gave us initial hit rates of 1.5% to 3.9%. We further filtered those hits to identify wells that only met our 60% inhibition cutoff on a single RGS target, to focus on compounds that would be most promising in terms of specificity. This filtering resulted in a hit rate between 0.3% and 1.4%, considerably lower and more amenable to follow-up experiments. Each of the hit wells was deconvoluted, as all 4 compounds in a hit well were cherrypicked for dose-response experiments. For the follow-up, dose-response confirmation rates were between 4% and 15%, somewhat lower than the expected 25%, representing 1 of 4 compounds in the mixture having activity, but not outside of a reasonable confirmation range. With 30 compounds exhibiting dose-response activity of 8,000 screened, our overall confirmed hit rate is 0.375%.
The revelation of CCG-50014 acting as an inhibitor of the RGS16/Gαo interaction indicates the potential of false-negative wells in this screening paradigm, and our overall confirmation rate of 4% to 15% reflects the presence of false-positive wells. In these cases, false-positives could stem from several sources, such as cross-compound reactivity, compound/protein aggregation or precipitation. It is unlikely that a spectral artifact of the combined compounds caused a well to be interpreted as a hit, due to the detection of fluorescence being gated on a bead-event. In addition, significant corruption of a bead's intrinsic spectral properties by a colored compound would result in a bead set being shifted outside of its tightly gated window on the Luminex, registering as a loss of events and not as a loss of bead-associated fluorescence. Every higher throughput method is subject to the generation of false positives and false negatives. The FCPIA method eliminates or reduces one of the major sources of false positives by virtually eliminating the effect of colored compounds in a well affecting the readout. In addition, our assay design of using two RGS family members and following up, using dose response experiments, on each family member helps to identify the presence of family-specific inhibitors and reveal those compounds in wells that may otherwise be false-negatives.
Perhaps most interesting is that of 24 wells with activity in the primary screen for which confirmed active compounds were found, 18 wells yielded a single compound while 6 wells yielded two actives for a total of 30 confirmed actives. This shows that we can identify compounds with activity from the 4-component mixtures using our polyplexed system.
Given the similarity between the 3 members of the RGS4 family in our screen (4,8,16), we followed up on each compound that had dose-response characteristics with each of the 3 family members. Several of the compounds that were identified as hits on one RGS has activity, albeit lower than our 60% cutoff, at 10μM, our approximate screening concentration. Interestingly, of the 25 compounds that had activity at one RGS among the R4 family members, 8 showed dose-response on at least one of the other members of the R4 family, as shown in Table 3.
In our previous experience, RGS4 is very sensitive to electrophonic compounds that may be able to covalently modify the cysteine residues on RGS4, resulting in a loss of activity. With this in mind, we created an RGS4 construct in which each of its 7 cysteine residues have been mutated to alanine. This “cysless” RGS4 binds Gαo with nearly identical affinity as the wild-type protein, and is not susceptible to the aforementioned inactivating modifications. When we re-tested 7 compounds that showed confirmatory dose-response activity on RGS4 (Figure 5A), at the cysless RGS4 constructs, each of them exhibited a marked loss of potency (Figure 5B). This indicates that our most potent lead compounds, CCG-50014 and CCG-55919, likely act through a mechanism that involves the modification of cysteine residues on RGS4. Interestingly, 3 compounds still showed some inhibition at the cysless RGS4 construct, but only at high concentrations (>30 to 100μM), where non-specific effects such as aggregation or other artifacts may become problematic.
In conclusion, we have demonstrated the feasibility of using polyplexed FCPIA as a method to detect inhibitors of protein-protein interactions. This study demonstrates the possibility of making multiple protein affinity measurements accurately within a single well, as well as the ability to deconvolute and identify single active compounds from complex mixtures. Using polyplexed FCPIA also provides an opportunity to have a series of concurrent counter-screens run in realtime with the high throughput screen. With proper data curation via a database (M-Screen), we are able to identify potential “family-wide” inhibitors, as well as wells containing compounds that inhibit all measured activity. The increased content available using polyplexing with a negligible time penalty greatly increases the utility of using flow cytometry as a method of high throughput screening for inhibitors of protein-protein interaction pairs.
ACKNOWLEDGEMENTS
The authors thank Levi L. Blazer and Donald J. Bern steel for assistance in the purification of several of the proteins used in this study. We also thank Martha J. Larsen and Renju T. Jacob of the Center for Chemical Genomics for the preparation of the screening mixture plates, cherrypicking and deconvolution. We thank Susan M. Wade for critical reading of the manuscript, and Dr. Larry Sklar (University of New Mexico) for his role developing and advancing the methods and technology for FCPIA.
Financial support of these studies included grants from the National Institutes of Health: NS057014 & DA023252 (RRN) and GM076821 (DLR) as well as the University of New Hampshire Biomolecular Interactions Technology Center (BITC) and an academic alliance with the Luminex Corporation. In addition, the cost of DNA sequencing was subsidized by the University of Michigan Comprehensive Cancer Center.
References
- (1).Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Molecular pharmacology. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- (2).Nieuwenhuijsen BW, Huang Y, Wang Y, Ramirez F, Kalgaonkar G, Young KH. A dual luciferase multiplexed high-throughput screening platform for protein-protein interactions. J Biomol Screen. 2003;8:676–684. doi: 10.1177/1087057103258287. [DOI] [PubMed] [Google Scholar]
- (3).Evelyn CR, Wade SM, Wang Q, Wu M, Iniguez-Lluhi JA, Merajver SD, Neubig RR. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6:2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- (4).Lan KL, Sarvazyan NA, Taussig R, Mackenzie RG, DiBello PR, Dohlman HG, Neubig RR. A point mutation in Galphao and Galphai1 blocks interaction with regulator of G protein signaling proteins. The Journal of biological chemistry. 1998;273:12794–12797. doi: 10.1074/jbc.273.21.12794. [DOI] [PubMed] [Google Scholar]
- (5).Sarvazyan NA, Remmers AE, Neubig RR. Determinants of gi1alpha and beta gamma binding. Measuring high affinity interactions in a lipid environment using flow cytometry. The Journal of biological chemistry. 1998;273:7934–7940. doi: 10.1074/jbc.273.14.7934. [DOI] [PubMed] [Google Scholar]
- (6).Sarvazyan NA, Lim WK, Neubig RR. Fluorescence analysis of receptor-G protein interactions in cell membranes. Biochemistry. 2002;41:12858–12867. doi: 10.1021/bi026212l. [DOI] [PubMed] [Google Scholar]
- (7).Sklar LA, Vilven J, Lynam E, Neldon D, Bennett TA, Prossnitz E. Solubilization and display of G protein-coupled receptors on beads for real-time fluorescence and flow cytometric analysis. BioTechniques. 2000;28:976–980. 982–975. doi: 10.2144/00285rr03. [DOI] [PubMed] [Google Scholar]
- (8).Sklar LA, Edwards BS, Graves SW, Nolan JP, Prossnitz ER. Flow cytometric analysis of ligand-receptor interactions and molecular assemblies. Annual review of biophysics and biomolecular structure. 2002;31:97–119. doi: 10.1146/annurev.biophys.31.082901.134406. [DOI] [PubMed] [Google Scholar]
- (9).Simons PC, Biggs SM, Waller A, Foutz T, Cimino DF, Guo Q, Neubig RR, Tang WJ, Prossnitz ER, Sklar LA. Real-time analysis of ternary complex on particles: direct evidence for partial agonism at the agonist-receptor-G protein complex assembly step of signal transduction. The Journal of biological chemistry. 2004;279:13514–13521. doi: 10.1074/jbc.M310306200. [DOI] [PubMed] [Google Scholar]
- (10).Hepler JR. Emerging roles for RGS proteins in cell signalling. Trends PharmacolSci. 1999;20:376–382. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- (11).Neubig RR. Regulators of G protein signaling (RGS proteins): novel central nervous system drug targets. J Pept Res. 2002;60:312–316. doi: 10.1034/j.1399-3011.2002.21064.x. [DOI] [PubMed] [Google Scholar]
- (12).Zhong H, Neubig RR. Regulator of G protein signaling proteins: novel multifunctional drug targets. The Journal of pharmacology and experimental therapeutics. 2001;297:837–845. [PubMed] [Google Scholar]
- (13).Wells J, Arkin M, Braisted A, DeLano W, McDowell B, Oslob J, Raimundo B, Randal M. Drug discovery at signaling interfaces. Ernst Schering Research Foundation workshop. 2003:19–27. doi: 10.1007/978-3-662-05314-0_3. [DOI] [PubMed] [Google Scholar]
- (14).Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nature reviews. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- (15).Pagliaro L, Felding J, Audouze K, Nielsen SJ, Terry RB, Krog-Jensen C, Butcher S. Emerging classes of protein-protein interaction inhibitors and new tools for their development. Current opinion in chemical biology. 2004;8:442–449. doi: 10.1016/j.cbpa.2004.06.006. [DOI] [PubMed] [Google Scholar]
- (16).Gilman AG. G proteins: transducers of receptor-generated signals. Annual review of biochemistry. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- (17).Navon SE, Fung BK. Characterization of transducin from bovine retinal rod outer segments. Mechanism and effects of cholera toxin-catalyzed ADP-ribosylation. The Journal of biological chemistry. 1984;259:6686–6693. [PubMed] [Google Scholar]
- (18).Breitwieser GE, Szabo G. Mechanism of muscarinic receptor-induced K+ channel activation as revealed by hydrolysis-resistant GTP analogues. The Journal of general physiology. 1988;91:469–493. doi: 10.1085/jgp.91.4.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mukhopadhyay S, Ross EM. Rapid GTP binding and hydrolysis by G(q) promoted by receptor and GTPase-activating proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9539–9544. doi: 10.1073/pnas.96.17.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lan KL, Zhong H, Nanamori M, Neubig RR. Rapid kinetics of regulator of G-protein signaling (RGS)-mediated Galphai and Galphao deactivation. Galpha specificity of RGS4 AND RGS7. The Journal of biological chemistry. 2000;275:33497–33503. doi: 10.1074/jbc.M005785200. [DOI] [PubMed] [Google Scholar]
- (21).Neubig RR, Connolly MP, Remmers AE. Rapid kinetics of G protein subunit association: a rate-limiting conformational change? FEBS letters. 1994;355:251–253. doi: 10.1016/0014-5793(94)01212-1. [DOI] [PubMed] [Google Scholar]
- (22).Traynor JR, Neubig RR. REGULATORs OF G PROTEIN SIGNALING & DRUGS OF ABUSE. Molecular interventions. 2005;5:30–41. doi: 10.1124/mi.5.1.7. [DOI] [PubMed] [Google Scholar]
- (23).Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nature reviews. 2002;1:187–197. [Google Scholar]
- (24).Roof RA, Jin Y, Roman DL, Sunahara RK, Ishii M, Mosberg HI, Neubig RR. Mechanism of action and structural requirements of constrained peptide inhibitors of RGS proteins. Chemical biology & drug design. 2006;67:266–274. doi: 10.1111/j.1747-0285.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- (25).Jin Y, Zhong H, Omnaas JR, Neubig RR, Mosberg HI. Structure-based design, synthesis, and activity of peptide inhibitors of RGS4 GAP activity. Methods in enzymology. 2004;389:266–277. doi: 10.1016/S0076-6879(04)89016-5. [DOI] [PubMed] [Google Scholar]
- (26).Young KH, Wang Y, Bender C, Ajit S, Ramirez F, Gilbert A, Nieuwenhuijsen BW. Yeast-based screening for inhibitors of RGS proteins. Methods in enzymology. 2004;389:277–301. doi: 10.1016/S0076-6879(04)89017-7. [DOI] [PubMed] [Google Scholar]
- (27).Fitzgerald K, Tertyshnikova S, Moore L, Bjerke L, Burley B, Cao J, Carroll P, Choy R, Doberstein S, Dubaquie Y, Franke Y, Kopczynski J, Korswagen H, Krystek SR, Lodge NJ, Plasterk R, Starrett J, Stouch T, Thalody G, Wayne H, van der Linden A, Zhang Y, Walker SG, Cockett M, Wardwell-Swanson J, Ross-Macdonald P, Kindt RM. Chemical genetics reveals an RGS/G-protein role in the action of a compound. PLoS genetics. 2006;2:e57. doi: 10.1371/journal.pgen.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lee E, Linder ME, Gilman AG. Expression of G-protein alpha subunits in Escherichia coli. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- (29).Sternweis PC, Robishaw JD. Isolation of two proteins with high affinity for guanine nucleotides from membranes of bovine brain. J Biol Chem. 1984;259:13806–13813. [PubMed] [Google Scholar]
- (30).Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- (31).Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D'Alecy LG, Neubig RR. Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol Cell Biol. 2006;26:6870–6879. doi: 10.1128/MCB.00314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]