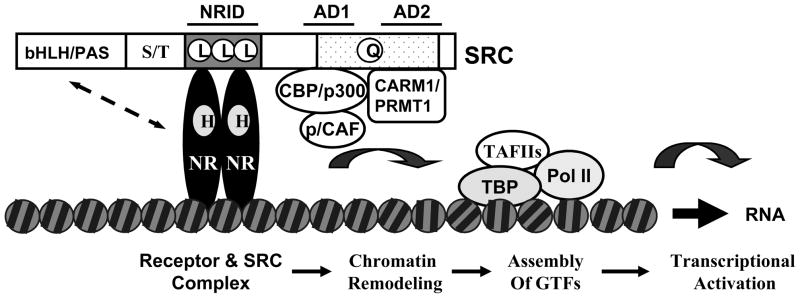
Fig. 1.
Molecular structure of SRCs and their functional mechanisms in steroid hormone-induced gene expression. The locations of basic structural and functional domains of SRCs are indicated. Upon hormone (H) binding, the hormone nuclear receptors (NR) expose their coactivator-binding motifs in their ligand-binding domains and allow SRCs to be recruited to the enhancer region of the NR target genes. SRCs further interact with CBP, p300, p/CAF, CARM1 and PRMT1 and recruit these common coactivators to the chromatin to build up a steroid receptor-directed transcriptional activation complex. This protein complex uses its protein acetyltransferase and methyltransferase activities to remodel the chromatin structure and to facilitate the assembly of general transcription factors and RNA polymerase II on the promoter for transcriptional activation. Of note, in addition to interactions between NR and the NRID domain of SRCs, interactions between NR and the bHLH/PAS domain of SRCs have been documented and may be important for function (see dotted line with arrowheads). Abbreviations: NRID, NR interaction domain; AD1 and AD2, activation domains 1 and 2; bHLH/PAS, the basis helix-loop-helix/Per-Ah receptor nuclear translocator-Sim domain; S/T, the serine and threonine-rich domain; L, L and L, the three LXXLL motifs responsible for interaction with nuclear receptors; Q, the glutamine-rich region; HAT, the histone acetyltransferase domain; CBP, the CREB (cAMP response element-binding protein) binding protein; p300, the 300 kDa protein homologous to CBP; p/CAF, the p300 and CBP-associated factor; CARM1, the coactivator-associated arginine methyltransferase 1; PRMT1, the protein arginine methyltransferase 1; TBP, the TATA binding protein; TAFIIs, TBP-associated general transcription factors (GTFs); Pol II, RNA polymerase II.