SUMMARY
To transit between hosts, intracellular Legionella pneumophila transform into a motile, infectious, transmissive state. Here we exploit the pathogen’s life cycle to examine how guanosine tetraphosphate (ppGpp) and DksA cooperate to govern bacterial differentiation. Transcriptional profiling revealed that during transmission alarmone accumulation increases the mRNA for flagellar and Type IV-secretion components, secreted host effectors, and regulators, and decreases transcripts for translation, membrane modification and ATP synthesis machinery. DksA is critical for differentiation, since mutants are defective for stationary phase survival, flagellar gene activation, lysosome avoidance, and macrophage cytotoxicity. The roles of ppGpp and DksA depend on the context. For macrophage transmission, ppGpp is essential, whereas DksA is dispensable, indicating ppGpp can act autonomously. In broth, DksA promotes differentiation when ppGpp levels increase, or during fatty acid stress, as judged by flaA expression and evasion of degradation by macrophages. For flagella morphogenesis, DksA is required for basal fliA (σ28) promoter activity. When alarmone levels increase, DksA cooperates with ppGpp to generate a pulse of Class II rod RNA or to amplify the Class III sigma factor and Class IV flagellin RNAs. Thus, DksA responds to the level of ppGpp and other stress signals to coordinate L. pneumophila differentiation.
Keywords: flagella, DksA, FliA, ppGpp, Legionella
INTRODUCTION
During infection, pathogens respond to local cues by altering their metabolism and virulence factor production. To coordinate physiological adaptation with virulence mechanisms, many pathogens employ two factors known to be critical for stationary phase resilience of most bacterial species, guanosine tetraphosphate (ppGpp) and DksA. Components of the stringent response regulate processes as diverse as Mycobacterium tuberculosis persistence (Dahl et al, 2003; Stallings et al, 2009), Salmonella enterica invasion (Pizarro-Cerda & Tedin, 2004; Thompson et al, 2006), Shigella flexneri intercellular spread (Sharma & Payne, 2006), and Legionella pneumophila transmission (Hammer & Swanson, 1999; Dalebroux et al, 2009).
L. pneumophila resides in aquatic reservoirs within biofilms or protozoa. When humans inhale contaminated aerosols, L. pneumophila infects alveolar macrophages. In host cells, the bacteria differentiate between two forms, replicative and transmissive (Molofsky & Swanson, 2004; Bruggemann et al, 2006) . Upon phagocytosis, transmissive bacteria utilize the Dot/Icm Type IV secretion system to avoid lysosomes and traffic to a vacuole derived from the endoplasmic reticulum (Isberg et al, 2009). In this compartment, bacteria that sense favorable conditions repress transmissive functions and activate genes needed for protein synthesis and replication (Sauer et al, 2005; Bruggemann et al, 2006). Eventually conditions deteriorate, cueing bacteria to synthesize ppGpp. The alarmone triggers differentiation to the motile, transmissive form, which resist degradation, lyse the exhausted host cell and are equipped to infect naive host cells (Molofsky & Swanson, 2004; Dalebroux et al, 2009). Under certain conditions, transmissive L. pneumophila develop into ‘mature intracellular forms’, which are fit to persist in the environment (Faulkner et al, 2008).
In most gamma-proteobacteria, including L. pneumophila, ppGpp levels are controlled by the synthetase RelA and the bifunctional synthetase/hydrolase SpoT. RelA monitors amino acid availability through its association with the ribosome, whereas SpoT responds to a variety of stimuli including fatty acid starvation, which requires direct interaction with acyl-carrier protein (Potrykus & Cashel, 2008; Battesti & Bouveret, 2009). Synthesis of ppGpp by exponential (E), replicative phase broth-grown L. pneumophila triggers differentiation to the post-exponential (PE), transmissive form (Hammer & Swanson, 1999; Dalebroux et al, 2009). For transmission between macrophages, ppGpp synthesized from SpoT is sufficient (Dalebroux et al, 2009). When conditions are favorable in host cells or in media, transmissive bacteria require SpoT to hydrolyze ppGpp and initiate replication. Thus, L. pneumophila modulates ppGpp levels to coordinate timely differentiation.
Many of the physiological effects of ppGpp are mediated through interactions with RNA polymerase (RNAP) in cooperation with the RNAP secondary channel interacting protein DksA (Haugen et al, 2008; Potrykus & Cashel, 2008). Whether ppGpp and DksA co-exert positive or negative regulation depends upon intrinsic properties of the promoters. While repressing ribosomal RNA operons (rRNA), ppGpp and DksA activate amino acid biosynthetic operons and alternative metabolic pathways. Direct co-positive regulation has also been observed during in vitro studies of promoters of critical virulence regulators (Nakanishi et al, 2006; Sharma & Payne, 2006; Aberg et al, 2008). Recently, DksA and ppGpp were shown to directly inhibit transcription of the E. coli σ70-dependent promoters of critical flagellar gene regulators, flhDC and fliA (σ28) to repress flagellar synthesis during starvation (Lemke et al, 2009).
Indirect transcriptional control reflects the impact of ppGpp and DksA on RNAP availability. During E. coli growth, nearly half the cellular RNAP is localized to rRNA operons by the σ70 vegetative sigma factor (Bremer, 1996). Upon nutrient limitation, ppGpp and DksA deactivate transcription from these loci, increasing the amount of core RNAP available to alternative sigma factors. These specialized subunits then direct polymerase to promoters of genes involved in particular stress responses (Bernardo et al, 2006; Szalewska-Palasz et al, 2007; Costanzo et al, 2008; Gummesson et al, 2009). Therefore, ppGpp and DksA indirectly control transcription by alternative sigma factors.
Recent evidence from E. coli suggests that DksA is more than a cofactor for ppGpp-dependent transcriptional control. Overproduction of DksA in ppGpp0 bacteria can compensate for lack of alarmone, indicating that DksA can act independently of ppGpp (Potrykus & Cashel, 2008). Additionally, phenotypic and in vitro assays show that ppGpp and DksA oppositely regulate some processes and promoters (Magnusson et al, 2007, Lyzen et al, 2009). Therefore, we exploited L. pneumophila differentiation and its flagellar cascade to investigate the functional relationship between ppGpp and DksA.
RESULTS
ppGpp induces rapid accumulation of virulence transcripts
Although ppGpp is known to induce L. pneumophila transmission traits, genes regulated by the alarmone have not been identified. To begin to define the ppGpp regulon, we developed a genetic system to synchronize ppGpp accumulation by L. pneumophila. When treated with IPTG, a relA spoT double mutant strain (ppGpp0) that carries an inducible allele of relA is locked in the transmissive state and exhibits heightened virulence (Dalebroux et al, 2009). Although replication is stunted, viability is not compromised. By 60 min after IPTG addition, a pool of ppGpp was evident in ppGpp0 prelAL.p. cell extracts, but not in ppGpp0 pempty controls; by 90 min, this pool had increased (Fig. 1).
Figure 1. Kinetics of ppGpp accumulation after prelAL.p induction.
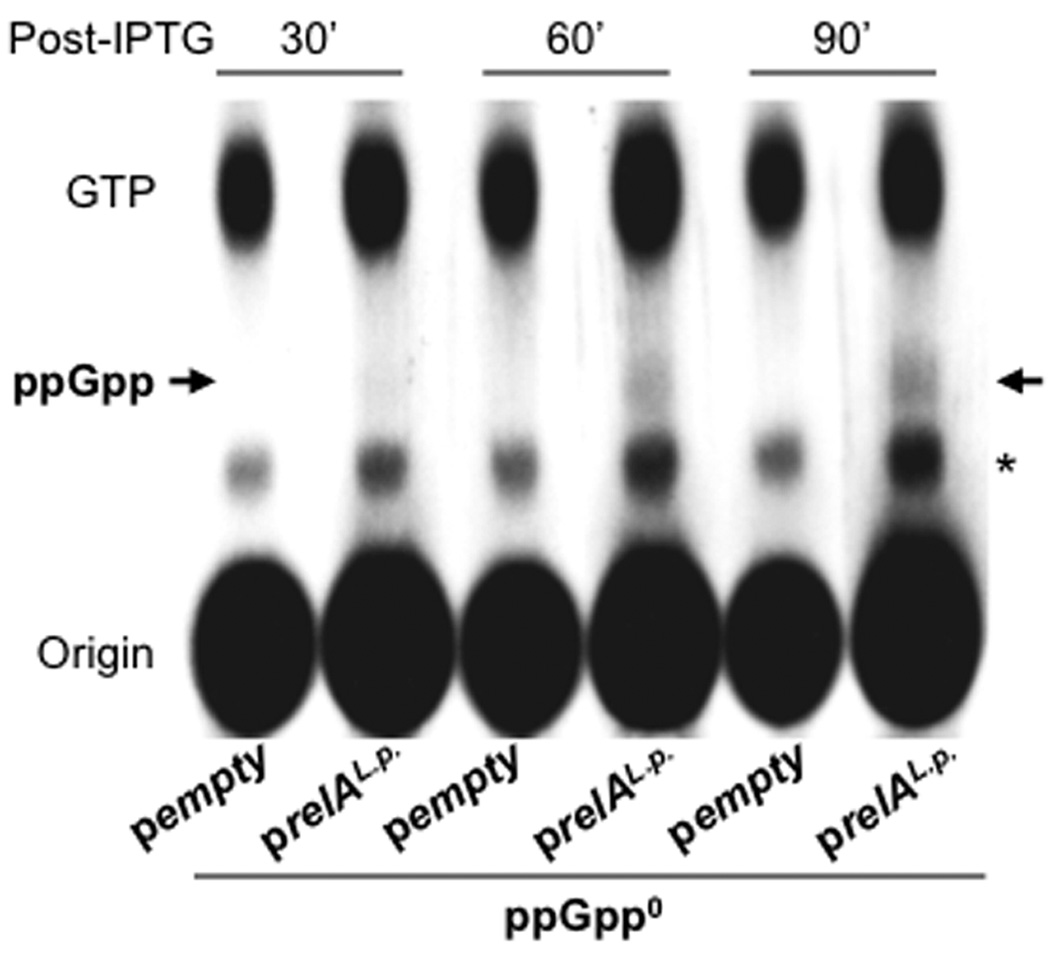
To evaluate the timing of ppGpp accumulation after relAL.p. induction, early-E phase AYET broth cultures of ppGpp0 pempty and ppGpp0 prelAL.p. labeled with 32P phosphoric acid were treated with 500 µM IPTG for the periods indicated, then nucleotides in cell extracts were separated by PEI-TLC. An arrow indicates the major ppGpp species, guanosine tetraphosphate; an asterisk denotes the position of guanosine pentaphosphate and an unidentified phosphorylated species. The autoradiogram shown represents one of two independent experiments.
To identify ppGpp-sensitive transcripts, cDNA prepared from ppGpp0 prelAL.p. and ppGpp0 pempty bacteria harvested at 45 and 90 min post-induction was hybridized to a comprehensive L. pneumophila microarray (Bruggemann et al, 2006), and relative transcript levels were calculated. This experimental design will identify loci whose expression is affected by cell growth, together with genes for which ppGpp accumulation activates or represses transcription initiation or transcript stability. For brevity, we will use “up-” and “down-regulation” to describe the relative differences between the two cultures, but kinetic studies of the corresponding promoters and transcripts will be required to identify the mechanism(s) that contribute.
Microarray results were validated by qRT-PCR analysis of six loci, and a strong correlation was apparent (R2 = 0.98; Fig. S1). Although statistically significant differences were observed at 45 min, most were less than two-fold, our criterion for ppGpp-regulated transcripts (http://www.ebi.ac.uk/microarray-as/ae/). Therefore, the 90 min data set was analyzed further. Many transcripts identified as ppGpp-sensitive at 90 min (Tables 1 and S1) were also differentially expressed at 45 min. By 90 min these differences had increased, suggesting that as time with ppGpp increased, existing differences were amplified.
Table 1.
Select list of transcripts up-regulated 90 min after relAL.p. induction.
| Gene Name / Function | Gene.ID | Annotation | ppGpp0 prelAL.p./pempty |
|---|---|---|---|
| Flagellar Genes | |||
| fleQ | lpg0853 | Master regulator of flagellar gene transcription (Class I) | 2.01 |
| fliH | lpg1758 | Export and assembly (Class IIa) | 2.41 |
| fliG | lpg1759 | Motor switch protein (Class IIa) | 3.88 |
| fleR | lpg1762 | Two-component response regulator (Class IIa) | 3.54 |
| fleS | lpg1763 | Two-component sensor histidine kinase (Class IIa) | 2.58 |
| fliN | lpg1791 | Motor switch protein (Class IIa) | 5.25 |
| flgB | lpg1216 | Proximal rod protein (Class IIb) | 4.32 |
| flgC | lpg1217 | Proximal rod protein (Class IIb) | 4.03 |
| flgG | lpg1221 | Distal rod protein (Class IIb) | 4.81 |
| flgH | lpg1222 | L-ring protein precursor (Class IIb) | 4.42 |
| flgJ | lpg1224 | Peptidoglycan-hydrolyzing protein (Class IIb) | 2.87 |
| fleN | lpg1783 | Regulator of flagellar synthesis (Class IIb) | 3.01 |
| flhF | lpg1784 | Biosynthesis regulator GTP-binding protein (Class IIb) | 3.87 |
| Dot/Icm Structural Genes | |||
| icmR | lpg0443 | Chaperone for IcmQ; a pore forming molecule | 2.46 |
| icmL/dotI | lpg0449 | Inner membrane protein | 2.23 |
| icmK/dotH | lpg0450 | Might be an outer membrane channel | 2.18 |
| icmE/dotG | lpg0451 | Major component of a channel | 1.96 |
| icmG/dotF | lpg0452 | Interacts with substrates; predicted channel component | 2.14 |
| icmC/dotE | lpg0453 | Predicted inner-membrane protein, Similar to DotV | 2.31 |
| icmD/dotP | lpg0454 | Predicted inner-membrane protein | 3.43 |
| icmJ/dotN | lpg0455 | Probable ATPase component | 2.10 |
| Dot/Icm Secreted Effectors | |||
| legC3 | lpg1701 | Coiled-coil domain, disrupts vacuolar trafficking, IcmSW-dependent | 2.11 |
| ylfA | lpg2298 | Coiled-coil domain, disrupts vacuolar trafficking, | 3.33 |
| sdeD | lpg2509 | Substrate of the Dot/Icm system | 1.98 |
| lepA | lpg2793 | SNAREs and coiled-coil domain; bacterial egress | 3.99 |
| Regulatory Factors | |||
| lpg0433 | lpg0433 | Putative transcriptional regulator | 1.99 |
| letE | lpg0537 | Transmission trait enhancer protein | 3.60 |
| rpoS | lpg1284 | RNA polymerase sigma factor (sigma-38) | 3.11 |
| relA | lpg1457 | ppGpp synthetase | 16.16 |
| lpg2732 | lpg2732 | Putative response regulator | 2.28 |
| rsmZ | non-coding, CsrA binding, regulatory RNA | 4.18 | |
| Known Virulence Factors | |||
| mip | lpg0789 | Macrophage infectivity potentiator | 2.06 |
| katB | lpg2389 | Catalase-peroxidase | 3.03 |
| enhC | lpg2639 | Enhanced entry protein | 2.73 |
| Fatty Acid / Carbon Metabolism | |||
| plsC | lpg0551 | similar to 1-acyl-sn-glycerol-3-phosphate acyltransferase | 2.51 |
| phaB1 | lpg0560 | Similar to acetoacetyl-CoA reductase | 2.09 |
| yfcX | lpg1596 | similar to alpha subunit of fatty-acid oxidation complex | 3.25 |
| phbC3 | lpg2260 | Similar to poly(3-hydroxyalkanoate) synthetase | 2.25 |
| pta | lpg2261 | Similar to phosphotransacetylase | 2.12 |
| pksJ | lpg2186 | Similar to polyketide synthase of type I | 2.17 |
| lpg2837 | lpg2837 | phospholipase/lecithinase/hemolysin, lysophospholipase A | 6.13 |
The data were collected from two independent biological replicates. See Fig. S1 for qRT-PCR validation. p < 0.01 for all targets shown.
Consistent with the E. coli stringent response to amino acid starvation (Durfee et al, 2008; Traxler et al, 2008), by 90 min of exposure to ppGpp, the transcripts for several ribosomal proteins were less abundant (Table S1). RNAs encoding components of the ATP synthase complex also decreased. Strikingly, RNAs for enzymes involved in lipopolysaccharide (LPS) modification and phospholipid biosynthesis were less abundant, including plsB, a known mediator of ppGpp-dependent regulation of fatty acid and phospholipid biosynthesis in E. coli (Heath et al, 1994). Thus, L. pneumophila employs ppGpp for stringent control over its membrane machinery and energy intensive processes like protein synthesis.
RNAs for several virulence factor loci accumulated within 90 min of relA induction. Consistent with their motility and toxicity toward macrophages (Dalebroux et al, 2009), ppGpp0 prelAL.p. bacteria contained significantly more transcripts for structural and regulatory elements of the flagellar apparatus (Table 1). Additionally, the amount of RNAs encoding membrane, channel and other components of the Dot/Icm Type IV secretion system increased. Consistent with transcriptional activation and secretion of Type IV effectors in PE phase (Nagai et al, 2002; Tiaden et al, 2007; Rasis & Segal, 2009), we observed an increase in the legC3, ylfA, sdeD and lepA Dot/Icm effector RNAs. Transcripts encoding virulence regulators also accumulated, including the alternative sigma factor rpoS (Bachman & Swanson, 2001; Hovel-Miner et al, 2009), the transmission trait enhancer letE (Bachman & Swanson, 2004), and the regulatory RNA rsmZ (Rasis & Segal, 2009; Sahr et al, 2009). In addition, the macrophage infectivity potentiator mip (Cianciotto & Fields, 1992), the catalase peroxidase katB (Bandyopadhyay et al, 2003), and the enhancer of macrophage uptake enhC (Liu et al, 2008) were up-regulated. Together these data demonstrate that L. pneumophila employs ppGpp to down-regulate protein synthesis machinery and up-regulate factors dedicated to survival and transmission in host cells.
L. pneumophila encodes a DksA homologue required for stationary phase morphogenesis and survival
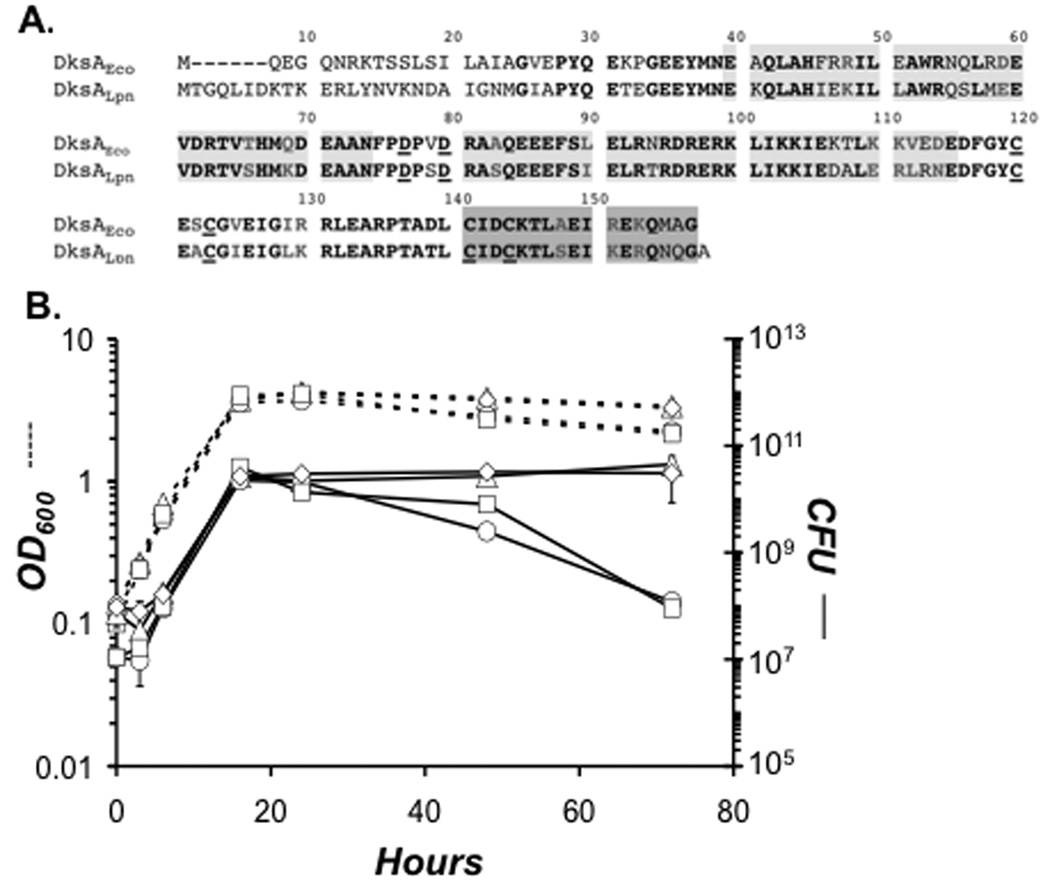
L. pneumophila encodes a 158 amino acid DksA-like protein (lpg2338) that is 72% identical to that of E. coli K-12 (Fig. 2A). Conserved in DksALpn are two aspartate residues known to comprise the acidic tip of DksAEco and other related RNAP secondary interacting proteins. The amino acid sequences in the two α-helical regions immediately adjacent to this pair of aspartates are also highly conserved, suggesting that DksALpn adopts a similar coiled-coil fold (Perederina et al, 2004). Also present in DksALpn are four cysteine residues known in DksAEco to comprise a zinc-finger motif. Obvious sequence dissimilarities lie at the extreme N-terminus, where the E. coli protein adopts a globular fold (Perederina et al, 2004). Therefore, L. pneumophila encodes a DksA protein with several but not all features of its E. coli counterpart.
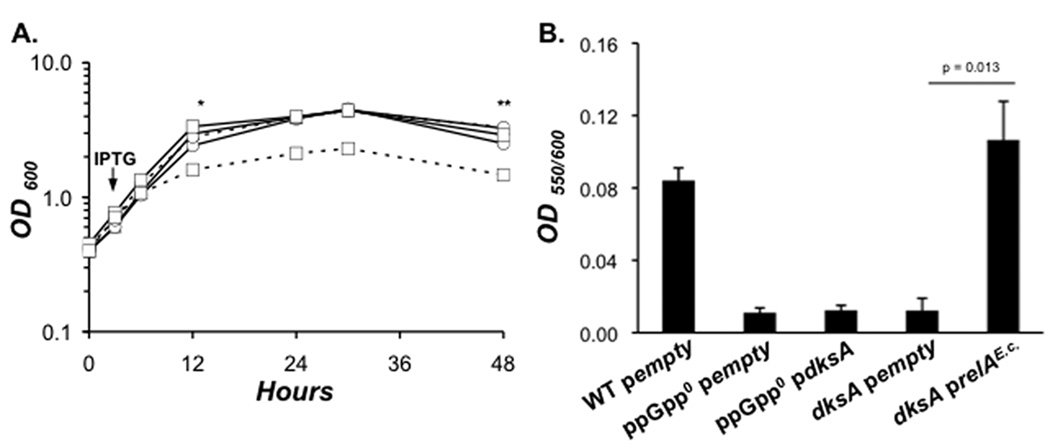
Figure 2. To survive stationary phase stress L. pneumophila requires DksA and ppGpp.
A. L. pneumophila encodes a 158 amino acid protein that is similar to the 151 amino acid DksA protein of E. coli K-12. Residues in bold, black lettering are identical; those in bold and grey are conserved. The underlined aspartate residues comprise the acidic tip, conserved in DksA and other RNAP secondary channel interacting proteins, while the underlined cysteine residues comprise a zinc finger motif. Light grey shading denotes two α-helical domains sandwiching the acidic tip that adopt a coiled-coil structure in the E. coli protein (Perederina et al, 2004). Dark grey shading indicates a C-terminal α-helix. B. Early-E phase AYET broth cultures of WT (triangles) and ppGpp0 (circles) carrying empty vector, and dksA mutants transformed with empty vector (squares) or pdksA (diamonds), were diluted to OD600 of 0.15, and bacteria were treated with 25 µM IPTG. At the times indicated, culture density and viability were quantified by reading OD600 (dashed lines) and enumerating colony forming units (CFU ± SE) ml−1 from duplicate samples on CYET (solid lines). The data represent one of three independent experiments.
To assess the contribution of DksA to differentiation, a dksA deletion insertion mutant was constructed and analyzed. Since L. pneumophila requires the ppGpp alarmone to survive in stationary phase (Dalebroux et al, 2009), we first tested if DksA also contributes to persistence in broth. From 3 – 24 h, both culture density and viability were similar for WT, ppGpp0, and dksA mutant bacteria (Fig. 2B). However, beginning in early stationary phase (~ 22 h), ppGpp0 and dksA mutant bacteria began to filament; by 48 h, or ~ 28 h into stationary phase, the mutants exhibited several defects. WT bacteria had become coccoid and motile (data not shown), two characteristics of transmissive cells (Molofsky & Swanson, 2004), whereas the ppGpp0 and dksA mutants remained amotile and had become more filamentous. Filamentation of dksA mutant bacteria was modest relative to ppGpp0 bacteria, which typically elongated to ~ 50 × the length of PE phase WT L. pneumophila (data not shown). After extended culture in stationary phase, ppGpp0 and dksA mutant bacteria lost viability: By 72 h, their CFU values had declined ~2 logs below the WT yield (Fig. 2B). The dksA mutant survival, morphogenesis, and motility defects were each restored when expression of plasmid-borne dksA was induced continuously from early-E phase to the late stationary phase with 25 µM isopropyl-beta-D-thiogalactopyranoside (IPTG; Fig. 2B; data not shown). Therefore, DksA and ppGpp each contribute to survival, morphology, and motility of stationary phase L. pneumophila.
To test the simple model that L. pneumophila require DksA to generate ppGpp, we performed thin layer chromatography. Similar to PE phase WT bacteria, dksA mutant L. pneumophila accumulated ppGpp, and the size of the pool increased with the length of the PE phase (data not shown). Therefore, the stationary phase defects of L. pneumophila dksA mutants could not be attributed to a deficiency in alarmone accumulation.
DksA contributes to PE phase virulence
Concomitant with its morphogenesis, stationary phase L. pneumophila activate several virulence phenotypes, including infectivity, lysosome evasion, cytotoxicity, and sensitivity to sodium, a phenotype associated with Type IV secretion (Vogel et al, 1996; Byrne & Swanson, 1998). Since ppGpp activated several loci implicated in particular transmisson traits, we investigated whether DksA also contributes.
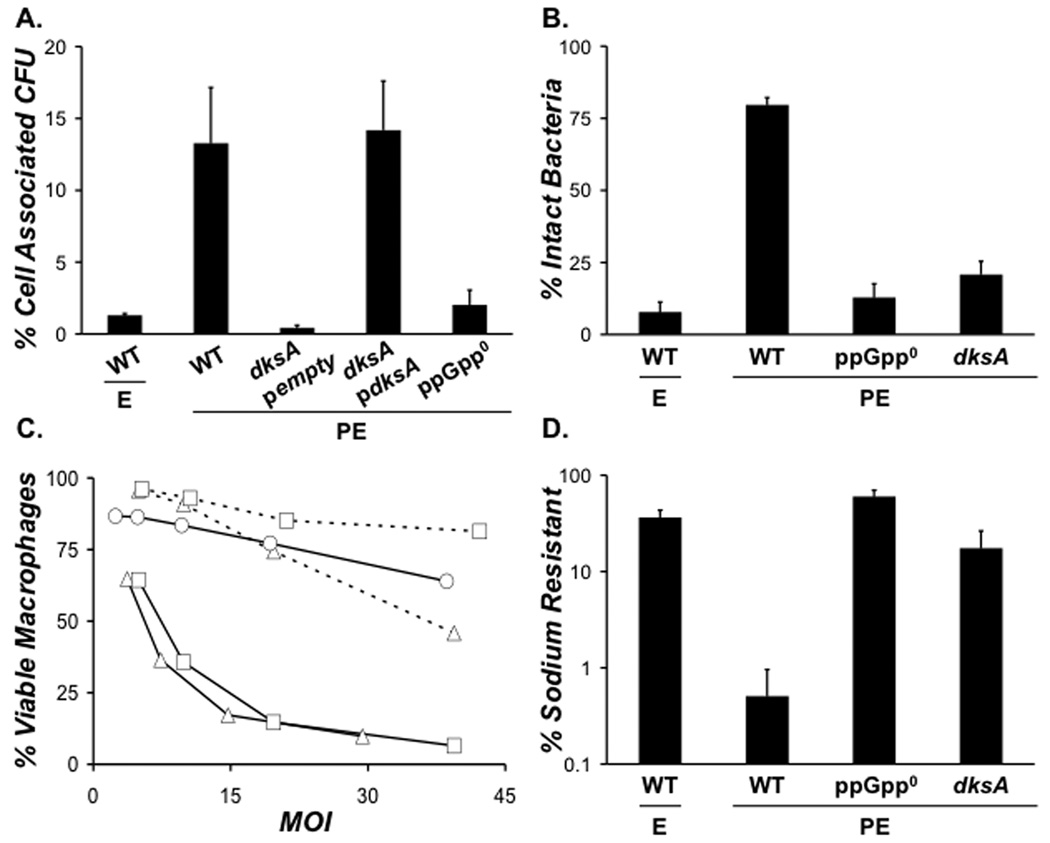
L. pneumophila required both DksA and ppGpp for PE phase activation of a panel of virulence phenotypes. Like E phase WT bacteria, PE phase dksA and ppGpp0 mutants were poorly infectious (Fig. 3A), frequently degraded within 2 h of infection (Fig. 3B), non-cytotoxic to macrophages (Fig. 3C), and sodium resistant (Fig. 3D). Induction of plasmid-borne dksA restored macrophage infectivity and cytotoxicity of the mutants (Fig. 3A and C). Thus, in broth, L. pneumophila employs both DksA and ppGpp to activate transmission traits.
Figure 3. To activate PE phase transmission traits L. pneumophila requires DksA and ppGpp.
A. To analyze whether DksA contributes to infectivity of PE phase L. pneumophila, macrophages were infected at an MOI of ~ 1 for 2 h with WT pempty, ppGpp0 pempty dksA pempty, or dksA pdksA cultured to the growth phase shown. Bacteria carrying plasmids were cultured from early-E phase to PE phase with 25 µM IPTG. Graphed are the mean percent of cell-associated CFU ± SE from duplicate wells in one of two independent experiments. B. The ability of bacteria to resist degradation in macrophage lysosomes was quantified using fluorescence microscopy by scoring the percent of intracellular bacteria that were intact at 2 h post-infection. Shown are the mean percentages from duplicate coverslips ± SE from three independent experiments. C. To determine the contribution of DksA to L. pneumophila cytotoxicity to macrophages, mid-E phase WT pempty (triangles, dashed lines), PE phase WT pempty (triangles, solid lines), PE phase ppGpp0 pempty (circles, solid lines), PE phase dksA pempty (squares, dashed lines), or PE phase dksA pdksA (squares, solid lines) bacteria, cultured with IPTG as described in A, were added to triplicate wells of macrophages at the MOI shown. The values plotted represent the mean ± SE for triplicate samples determined in one of three similar experiments. D. To measure sodium resistance, E or PE phase bacteria of the strains depicted were plated onto medium with or without 100 mM NaCl. Shown are the mean percentages from duplicate samples ± SE from three independent experiments.
DksA and ppGpp promote growth in amoebae
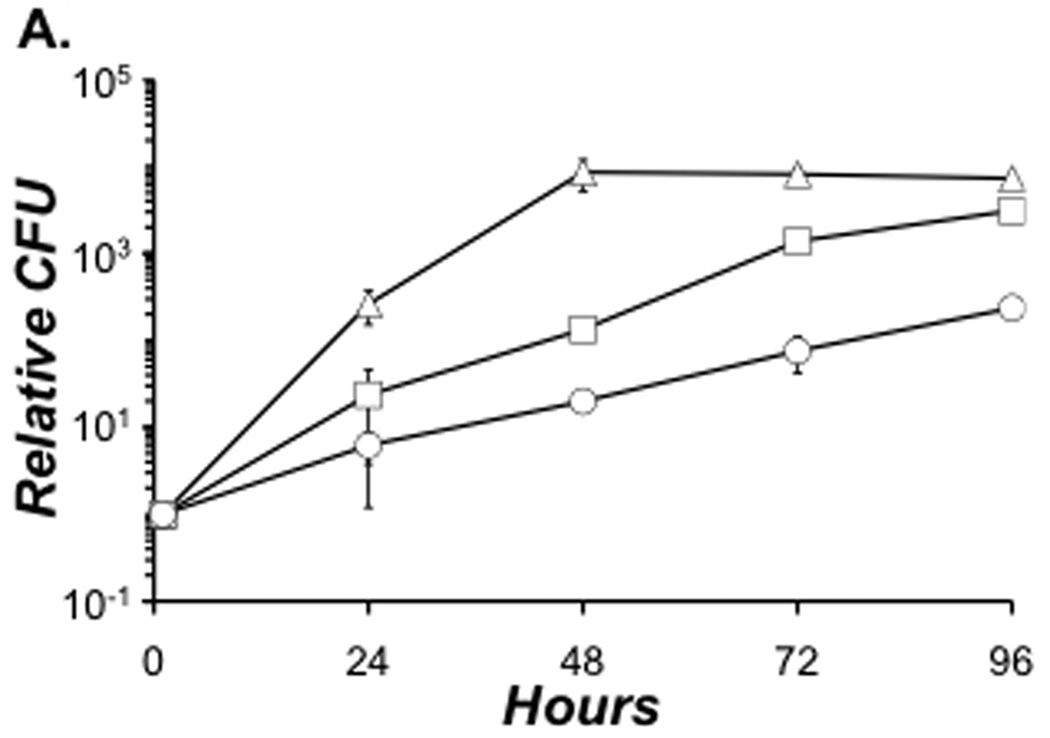
In the environment, L. pneumophila parasitizes numerous species of protozoa, including Acanthamoeba castellanii. Optimal growth in A. castellanii required both DksA and ppGpp. During the 4-day incubation, WT bacteria replicated robustly, increasing their CFU ~ 104 fold by 48 h and exhibiting a growth rate constant (µ) of 0.19 h−1 between 1 and 48 h (Fig. 4). The yield of dksA and ppGpp0 mutants was less than that of WT at each time analyzed (Fig. 4). Bacteria that lacked dksA were better equipped for intracellular replication than those that could not generate ppGpp (µ = 0.10 h−1 and 0.06 h−1, respectively, between 1 and 48 h). By 96 h, the yield of dksA CFU approached the WT level, while ppGpp0 mutant values were 1–2 logs lower (Fig. 4). Thus, for optimal growth in A. castellanii, L. pneumophila requires both DksA and ppGpp, but the alarmone is more critical.
Figure 4. For optimal growth in A. castellanii L. pneumophila requires both DksA and ppGpp.
Thy+ derivatives of WT (triangles), dksA (squares) and ppGpp0 (circles) bacteria were cultured to PE phase, then added to amoebae in infection buffer at an MOI of ~ 0.05. At the times shown aliquots were suspended and lysed, and lysates plated for CFU enumeration. To obtain relative CFU, viable counts for 24, 48, 72, and 96 were divided by the 1 h CFU value for each strain. Depicted are the relative CFU values from duplicate infections ± SE. The data represent one of two independent experiments.
L. pneumophila requires ppGpp but not DksA for transmission between macrophages
As in broth, L. pneumophila alternates between replicative and transmissive forms in amoebae and macrophages (Byrne & Swanson, 1998; Hammer & Swanson, 1999; Sturgill-Koszycki & Swanson, 2000; Alli et al, 2000; Molofsky & Swanson, 2003; Sauer et al, 2005; Bruggemann et al, 2006). Whereas ppGpp is dispensable for intracellular replication, the alarmone is essential for intracellular progeny to survive transmission from one macrophage to another (Dalebroux et al, 2009). Therefore, we analyzed the contribution of DksA to transmission in murine macrophages.
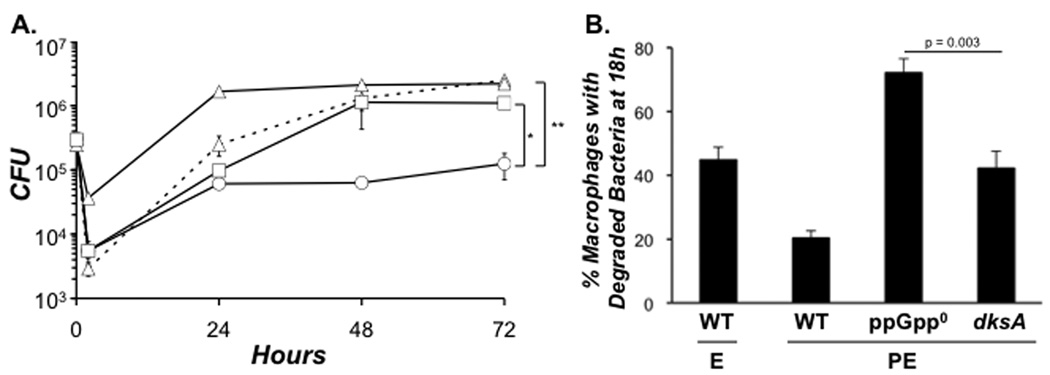
As reported in Figure 3A, DksA and ppGpp were essential for efficient infection by PE phase L. pneumophila. Both mutants behaved like E phase WT bacteria, yielding less CFU at 2 h than PE phase WT (Fig. 5A). As expected, neither DksA nor ppGpp were required during the primary replication period: From 2 – 24 h, their growth curves were nearly indistinguishable, each exhibiting an ~ 1 log increase in CFU. Between 18 and 24 h, when replicating WT bacteria differentiate to the transmissive form (Byrne & Swanson, 1998; Hammer & Swanson, 1999; Molofsky & Swanson, 2003), DksA was dispensable, but ppGpp was not. The dksA mutant CFU continued to increase until 48 h, when their numbers were similar to WT. In contrast, the yield of ppGpp0 mutants failed to increase throughout the remainder of the infection (Fig. 5A).
Figure 5. L. pneumophila requires ppGpp for macrophage transmission; DksA is less critical.
A. Macrophages were infected at an MOI of ~ 1 with E phase WT (triangles, dashed lines), PE phase WT (triangles, solid line), ppGpp0 (circles), or dksA (squares) bacteria. The number of viable bacteria was determined at the time points shown. Depicted are the mean CFU ± SE from duplicate samples in one of four independent experiments. * Indicates that the difference in the mean CFU values calculated from four independent experiments for WT and ppGpp0 mutant bacteria at 72 h was statistically significant by a paired, two-tailed Student t test (p = 0.047). ** Indicates that the difference between ppGpp0 and dksA mutant bacteria at 72 h was also statistically significant (p = 0.047). B. Macrophages were infected at an MOI of ~ 1 with PE phase WT or an MOI ~ 3 of E phase WT, PE phase ppGpp0 and PE phase dksA mutant bacteria, then at 18 h coverslips were fixed. A total of 100 infected macrophages from duplicate coverslips were scored as follows: macrophages with a single intact or a single degraded bacterium, and macrophages with multiple intact or multiple degraded bacteria. Shown is the mean percent ± SE of macrophages with degraded bacteria at 18 h from three independent experiments.
At the end of the primary replication period in macrophages, L. pneumophila that lack ppGpp are vulnerable to lysosomal degradation (Dalebroux et al, 2009). Consistent with their lower yield between 48 and 72 h, ppGpp0 mutants were more frequently degraded at 18 h than were dksA mutants, as judged by immunofluorescence microscopy (Fig. 5B). To assess the ability of transmitted L. pneumophila to resist lysosomal degradation, macrophages that contained only a single intact or degraded bacterium at 24 h were scored in two independent experiments. By this criterion, ppGpp0 mutant L. pneumophila were more susceptible to lysosomal degradation during host-to-host transmission than either dksA mutant or WT L. pneumophila, since 73% ± 8% of infected macrophages harbored a single degraded ppGpp0 bacterium, whereas 53% ± 5%, 51% ± 5%, and 49% ± 1% of infected macrophages contained a single degraded WT E, WT PE, or dksA bacterium, respectively. Together, the CFU and morphological data indicate that, whereas DksA is dispensable, L. pneumophila requires the alarmone ppGpp for efficient transmission to a new macrophage.
Constitutive ppGpp synthesis by L. pneumophila can halt replication and stimulate pigmentation independently of DksA
The distinct phenotypes of dksA and ppGpp0 mutant bacteria during growth in A. castellanii (Fig. 4) and transmission in macrophages (Fig. 5) motivated us to investigate whether ppGpp or DksA can activate L. pneumophila transmission traits independently of each other. For this purpose, dksA mutant L. pneumophila were transformed with prelAE.c., which encodes an inducible E. coli K-12 RelA with a C-terminal truncation that prevents its interaction with the ribosome, resulting in constitutive synthetase activity (Schreiber et al, 1991; Hammer & Swanson, 1999). In parallel, ppGpp0 bacteria were transformed with pdksA.
Induction of dksA did not impact replication of E phase L. pneumophila, since the culture densities of WT, ppGpp0 pempty, and ppGpp0 pdksA bacteria were similar after IPTG addition (Fig. 6A). In contrast, replication of dksA prelAE.c. cells halted shortly after IPTG addition, and their OD600 values subsequently remained lower than WT and dksA pempty control bacteria (Fig. 6A). Therefore, when expression of relAE.c. is highly induced, ppGpp can arrest the L. pneumophila cell cycle independently of DksA.
Figure 6. Constitutive ppGpp synthesis is sufficient to bypass the dksA requirement for cell cycle arrest and pigment production.
A. To test the effect of either dksA induction or constitutive ppGpp synthesis on the growth of L. pneumophila mutant bacteria, WT pempty (triangles), ppGpp0 transformed with either pempty (circles, solid lines), or pdksA (circles, dashed lines), and dksA mutants carrying either pempty (squares, solid lines), or prelAE.c. (squares, dashed lines), a truncated and constitutively active form of E. coli RelA, were induced with IPTG in early-E phase AYET broth cultures, then culture density was monitored by reading OD600 over the time period shown. B. At 48 h post-IPTG (indicated in A by **), the OD550 of the supernatants was read to quantify extracellular pigment production. To account for differences in cell density, pigmentation values were normalized to culture density by dividing OD550 by OD600. The graph depicts the mean OD550/OD600 values ± SE from three independent experiments.
In late stationary phase, L. pneumophila produce a secreted pyomelanin pigment by a pathway induced by RelA (Zusman et al, 2002; Chatfield & Cianciotto, 2007). Pigmentation required both DksA and ppGpp, as pyomelanin was not detected after a ~ 44 h treatment of dksA and ppGpp0 mutant bacteria that carried only the vector (Fig. 6B). Nevertheless, DksA was not essential to this response, since ectopic expression of relAE.c. by dksA mutants was sufficient to generate as much pyomelanin as WT cultures. Nor could DksA function independently of ppGpp to activate this pathway, since ppGpp0 pdksA cultures failed to pigment. Therefore, ppGpp can bypass the dksA requirement for pigment production, but the opposite is not true.
DksA controls the propionic acid response cooperatively and independently of ppGpp
When treated with 10 mM propionic acid, E phase L. pneumophila rapidly transition to the transmissive state, a response that is specific to carboxylic acids rather than pH (Edwards et al, 2009). To assess the contribution of DksA to L. pneumophila’s stringent response to fatty acid addition, we used a transcriptional reporter of transmission. The plasmid pflaAgfp encodes the flaA promoter fused to the gene encoding green fluorescent protein (gfp; Hammer & Swanson, 1999).
Neither ppGpp nor DksA is required for growth inhibition by propionic acid, since WT, ppGpp0 and dksA mutant cultures responded similarly (Fig. 7A, top). However, unlike WT bacteria, which exhibited heightened fluorescence 3 h post-propionic acid treatment, dksA mutant bacteria failed to fluoresce throughout the time course analyzed (Fig. 7A, bottom). Consistent with their lack of motility in the PE phase, dksA mutant control cultures did not activate flaA expression even when the cells reached stationary phase, while WT bacteria did (9 h; Fig. 7A, bottom). Therefore, L. pneumophila require DksA to induce the flaA promoter when the stringent response is triggered by propionic acid or at the transition to PE phase (Hammer & Swanson, 1999; Edwards et al, 2009).
Figure 7. In response to fatty acid stress, DksA activates transmission traits with and independently of ppGpp.
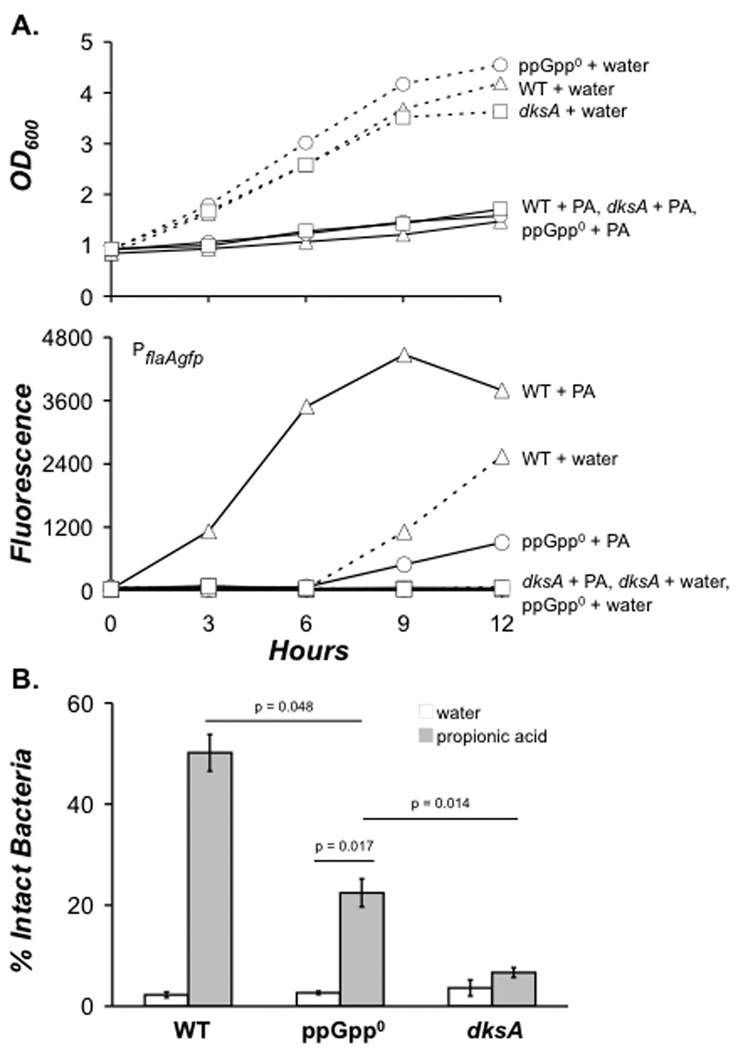
A. To determine whether DksA is required for flaA promoter activation in response to excess propionic acid (PA), mid-E phase bacteria harboring the pflaAgfp reporter plasmid were normalized to an OD600 (0.75–0.85), then 10 mM (PA) (solid lines), or water (dashed lines) were added to WT (triangles), ppGpp0 (circles), or dksA (squares) bacteria. At 3 h time intervals, cell density (top panel) and flaA promoter activity was quantified by measuring green fluorescent protein (gfp) accumulation using fluorometry. The data depicted represent one of three independent experiments. B. To determine if DksA contributes to increased resistance to degradation by macrophages after PA treatment, PA (light gray), or water (white) was added to cultures as described in A. After 3 h, bacteria were used to infect macrophages, and resistance to lysosomal degradation was scored by fluorescence microscopy. Shown are the mean percentages from triplicate coverslips ± SE from three independent experiments. In addition to the p values depicted, the mean percent resistance values for WT PA and dksA PA were statistically different by a two-tailed paired Student t-test (p = 0.005).
To verify its role in the L. pneumophila response to fatty acid perturbation, we analyzed the contribution of DksA to evasion of lysosomal degradation. After a 3 h propionic acid treatment, ~ 50% of the infected macrophages contained intact WT bacteria, whereas < 10% harbored intact dksA mutant bacteria (Fig. 7B). Therefore, L. pneumophila also require DksA to induce resistance to macrophage degradation when the stringent response is triggered by propionic acid.
Unlike DksA, ppGpp was only partially required for activation of the flaA-gfp transmission reporter. We repeatedly observed modest promoter activation 6 – 9 h after propionic acid addition to ppGpp0 cultures (Fig. 7A, bottom). In addition, ~ 20% of alarmone deficient bacteria avoided degradation, another intermediate response (Fig. 7B). Importantly, DksA accounted for the ppGpp-independent flaA promoter activity and degradation evasion, since dksA mutants treated with propionic acid did not increase their fluorescence (Fig. 7A, bottom) or remain intact in macrophages (Fig. 7B). Thus, L. pneumophila transmission trait activation in response to excess fatty acids is mediated by a DksA activity that is enhanced by ppGpp.
DksA induction leads to ppGpp-independent flagellar biosynthesis
To continue to investigate the capacity of DksA to act independently, we asked whether this transcription factor could control flagellar biogenesis in the absence of ppGpp. Using the strains characterized in Figure 6, we measured whether induction of dksA expression could rescue the flagellar synthesis defect of ppGpp0 mutant L. pneumophila.
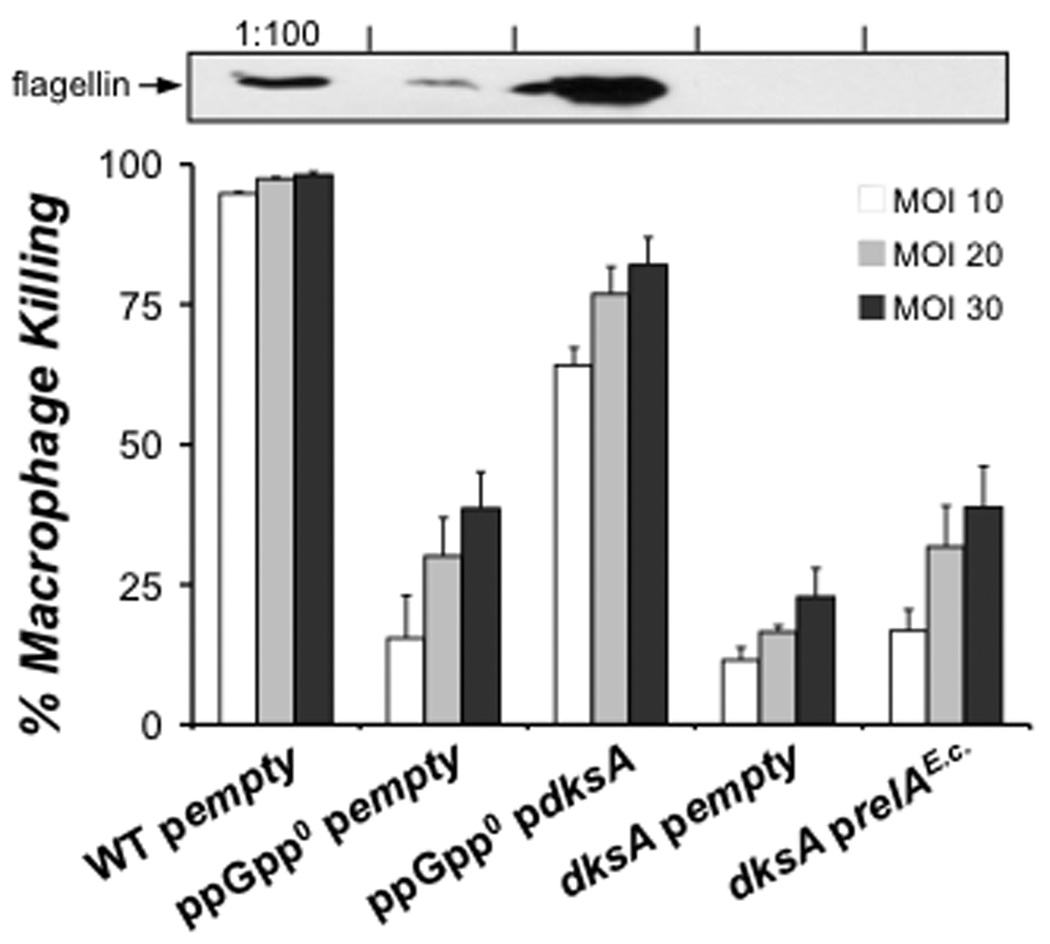
Once control bacteria entered stationary phase (~ 9 h post-IPTG; Fig. 6A), we assayed levels of flagellin and macrophage cytotoxicity, a flagellin-dependent phenotype (Molofsky et al, 2006). Induction of plasmid-borne dksA increased both the cell-associated flagellin and cytotoxicity of ppGpp0 bacteria (Fig. 8). Likewise, dksA induction restored motility of ~ 20 – 35% of the ppGpp0 bacteria. Full bypass of ppGpp by DksA was not achieved, as flagellin levels, macrophage killing and motility of ppGpp0 mutant bacteria were still lower than PE phase WT (Fig. 8). In contrast, relAE.c. induction did not restore flagellin synthesis, cytotoxicity or motility to dksA mutants (Fig. 8; data not shown). Therefore, when expressed from a plasmid, dksA can activate flagellin-dependent phenotypes independently of ppGpp. Furthermore, this contribution by DksA cannot be bypassed by constitutive ppGpp synthesis.
Figure 8. Induction of dksA is sufficient to bypass the requirement for ppGpp for flagellin-dependent phenotypes.
In the strains subjected to the induction described in Figure 6A, we assessed levels of flagellin and macrophage cytotoxicity. At 9 h post-IPTG (indicated in Fig. 6A by *), bacterial cell pellets were harvested, and levels of cell-associated flagellin were assessed by Western analysis (upper image). At 9 h post-IPTG, bacteria were also added to triplicate wells of macrophages at the MOIs shown (bottom panel), and cytotoxicity was measured. The values plotted represent the mean percent macrophage killing ± SE for triplicate samples determined in one of three similar experiments.
DksA contributes to the flagellar regulon
As an independent approach to analyze the contribution by DksA to the flagellar gene regulon, the transcriptional profiles of PE phase WT and dksA mutant L. pneumophila were compared (Table 2). Consistent with a role for DksA in the down-regulation of ribosomal genes in PE phase, several ribosomal transcripts were elevated in dksA mutants relative to WT bacteria (http://www.ebi.ac.uk/microarray-as/ae/). As in P. aeruginosa, L. pneumophila flagellar gene transcription occurs in a hierarchy of four classes (Dasgupta et al, 2003; Bruggemann et al, 2006; Heuner, 2007; Albert-Weissenberger et al, 2010). DksA was especially important for PE phase accumulation of late flagellar transcripts, including those encoding structural elements critical for final assembly and motor components essential for flagellar rotation (Table 2). In addition, Class II rod and hook transcripts and the Class III sigma factor fliA as well as each of its downstream flagellar gene targets required DksA for up-regulation (Table 2; Albert-Weissenberger et al, 2010). DksA also contributed to an increase in RNA for RpoN (σ54), a factor shown to play a modest role in PE phase up-regulation of a few Class II flagellar genes, including flgB (Albert-Weissenberger et al, 2010). Thus, L. pneumophila employs the DksA protein to increase the level of flagellar transcripts in the stationary phase of growth.
Table 2.
Flagellar gene expression of PE phase dksA mutant L. pneumophila.
| Gene Name | Gene.ID | Annotation | dksA/WT |
|---|---|---|---|
| Class I | |||
| rpoN | lpg0477 | RNA polymerase sigma factor (sigma-54) | −2.17 |
| fleQ | lpg0853 | Master regulator of flagellar gene transcription | --- |
| Class IIa | |||
| fliJ | lpg1756 | Export and assembly | --- |
| fliI | lpg1757 | Flagellum-specific ATP synthase | --- |
| fliH | lpg1758 | Export and assembly | --- |
| fliG | lpg1759 | Motor switch protein | --- |
| fliF | lpg1760 | M-ring protein | −2.17 |
| fliE | lpg1761 | Hook-basal body complex protein | --- |
| fleR | lpg1762 | Two-component response regulator | --- |
| fleS | lpg1763 | Two-component sensor histidine kinase | --- |
| flhA | lpg1785 | Export and assembly | --- |
| flhB | lpg1786 | Export and assembly | --- |
| fliR | lpg1787 | Export and assembly | --- |
| fliQ | lpg1788 | Export and assembly | --- |
| fliP | lpg1789 | Export and assembly | --- |
| fliO | lpg1790 | Export and assembly | --- |
| fliN | lpg1791 | Motor switch protein | −2.85 |
| fliM | lpg1792 | Motor switch protein | --- |
| Class IIb | |||
| flgA | lpg0908 | P-ring biosynthesis | --- |
| flgB | lpg1216 | Proximal rod protein | −2.38 |
| flgC | lpg1217 | Proximal rod protein | −4.55 |
| flgD | lpg1218 | Rod modification protein | −4.00 |
| flgE | lpg1219 | Flagellar hook protein | −11.11 |
| flgF | lpg1220 | Proximal rod protein | −4.34 |
| flgG | lpg1221 | Distal rod protein | --- |
| flgH | lpg1222 | L-ring protein precursor | --- |
| flgI | lpg1223 | P-ring protein precursor | --- |
| flgJ | lpg1224 | Peptidoglycan-hydrolyzing protein | --- |
| flgK | lpg1225 | Hook-associated protein | −2.70 |
| flgL* | lpg1226 | Flagellar hook-associated protein | −12.50 |
| fliK | lpg1688 | Hook-length control protein | −3.57 |
| fleN | lpg1783 | Regulator of flagellar synthesis | −7.14 |
| flhF | lpg1784 | Biosynthesis regulator GTP-binding protein | −6.25 |
| Class III | |||
| flgN* | lpg0906 | Potential chaperone | −8.33 |
| flgM* | lpg0907 | Anti-sigma-28 factor | −20.00 |
| motB* | lpg1780 | Sodium-type motor protein | −16.67 |
| motA* | lpg1781 | Sodium-type motor protein | −20.00 |
| fliA | lpg1782 | RNA polymerase sigma factor (sigma-28) | −20.00 |
| motA2 | lpg2318 | Proton conductor component of motor | −10.00 |
| motB2 | lpg2319 | Motor protein | −16.67 |
| flhB’* | lpg2583 | Unknown | −10.00 |
| Class IV | |||
| fliS* | lpg1337 | Potential chaperone | −20.00 |
| fliD* | lpg1338 | Filament cap | −25.00 |
| flaG* | lpg1339 | Unknown | −33.33 |
| flaA* | lpg1340 | Flagellin | −50.00 |
| motY* | lpg2962 | Sodium-type motor protein | −7.14 |
The data were collected from two independent biological replicates. See Fig. S2 for RT-PCR validation.
p < 0.01 for all targets shown.
indicates a gene regulated by fliA (σ28) (Albert-Weissenberger et al, 2010)
ppGpp modulates DksA-dependent expression of flagellar genes
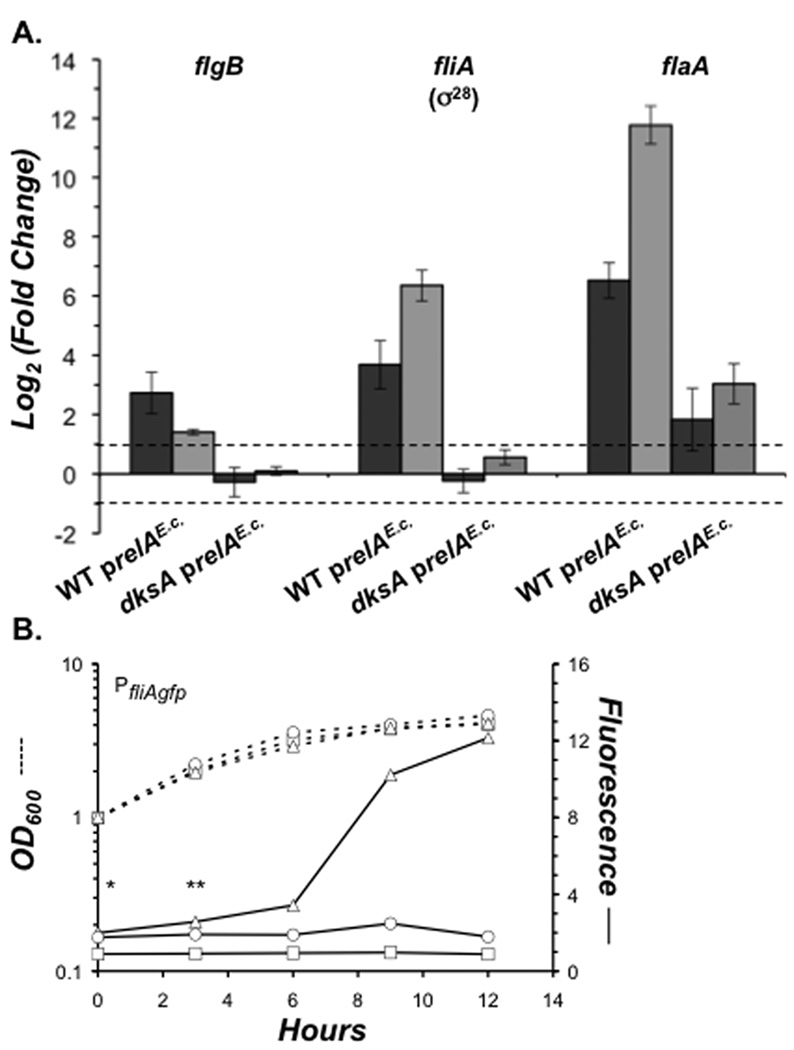
To investigate how DksA contributes to expression of flagellar genes stimulated by alarmone, we induced synchronous ppGpp synthesis by WT and dksA mutant bacteria, and then analyzed expression of genes representing three tiers of the flagellar regulon 0, 90 and 300 min later. By 300 min, 95–100% of WT cells were motile, verifying functional flagella. Furthermore, the growth of both WT and dksA mutant L. pneumophila was rapidly inhibited (Fig. S3), suggesting that the strains accumulated ppGpp similarly (Schreiber et al, 1991). Therefore, the fold-increase relative to t = 0 was calculated for three representative transcripts: flgB, a Class II gene encoding a flagellar basal-body rod protein; fliA, a putative a Class III gene encoding σ28, the sigma factor that activates flaA (Heuner et al, 2002); and flaA, a Class IV gene encoding flagellin.
By 90 min, flgB, fliA and flaA RNA levels had increased in WT bacteria, revealing that soon after ppGpp synthesis, each tier of the flagellar gene cascade is active (Fig. 9A). Between 90 and 300 min, fliA and flaA levels increased further in WT bacteria, whereas flgB levels declined. Therefore, as alarmone accumulates (Hammer & Swanson, 1999), expression of the Class III fliA and Class IV flaA loci is continuously induced, whereas expression of the Class II flgB gene is induced, and then repressed.
Figure 9. L. pneumophila uses ppGpp to modulate DksA-dependent gene expression.
A. To assess the contribution of DksA to gene expression by ppGpp, qRT-PCR was performed on transcripts isolated from mid-E phase AYET broth cultures of WT and dksA mutant bacteria induced to express prelAE.c. with 500 µM IPTG. RNA was harvested at 0, 90 and 300 minutes post-IPTG, and the relative transcript levels were assessed by dividing 90 min (dark grey) and 300 min (light grey) values by the 0 min value to give the fold increase for each target. Depicted are the mean log2 (fold increase) values from triplicate wells in three independent experiments ± SE. Dashed lines delineate the two-fold change cutoff. B. To monitor fliA promoter activity at the E to PE phase transition, mid-E phase WT (triangles), ppGpp0 (circles), or dksA (squares) bacteria harboring the pfliAgfp reporter plasmid were diluted to an OD600 of 1.0. Cell density (dashed lines) and fliA promoter activity (solid lines) were quantified until bacteria entered stationary phase. Actual fluorescence values were 1000× greater than those shown. The data represent one of three independent experiments, and the differences between WT and dksA, and ppGpp0 and dksA at 0* and 3 h** were statistically significant by a two-tailed paired Student t-test (p = 0.01*, 0.04** and 0.03*, 0.006**).
Rapid ppGpp-mediated up-regulation required DksA, as no increase was observed for these loci at 90 min post-induction in dksA mutants (Fig. 9A). The DksA contribution to flaA RNA accumulation could be partially bypassed by relAE.c. induction, as dksA mutants showed modest ~ 3-fold increase in flaA transcript at 300 min. However, when their flaA levels were compared directly, dksA mutants contained ~ 140 fold less flaA mRNA than WT bacteria (data not shown).
Consistent with fliA mRNA accumulation patterns, ppGpp and DksA were both required for activation of the fliA promoter at the E to PE phase transition, as judged by a fliA-gfp reporter (Fig. 9B). Additionally, DksA contributed to basal fliA promoter activity in E phase, since WT and ppGpp0 mutant fluorescence values were consistently 2–3 fold higher than those of dksA bacteria. Likewise, in the uninduced qRT-PCR samples, WT bacteria contained ~ 3 fold more fliA mRNA than dksA mutants (data not shown). Therefore, L. pneumophila requires the DksA transcription factor to activate the flagellar regulon, whereas the alarmone modulates this activation, by either amplifying or repressing flagellar gene expression over time.
DISCUSSION
When their capacity to synthesize either proteins or fatty acids is compromised, replicating L. pneumophila rely on the signaling molecule ppGpp to orchestrate differentiation to a transmissive, infectious form (Edwards et al, 2009; Dalebroux et al, 2009). In the process, intracellular L. pneumophila alter the level of ~ 800 transcripts (Bruggemann et al, 2006). Here we demonstrate that ppGpp and the transcription factor DksA can either cooperate or act independently to coordinate this developmental program.
Composed of over 40 genes whose expression occurs in four distinct phases, the flagellar transcriptional cascade provides a sensitive read-out for analyzing the impact of ppGpp and DksA during L. pneumophila differentiation. Several factors regulate the hierarchy, including the master regulator FleQ (Class II and III), the response regulator FleR and the alternative sigma factors σ54 (Class II) and σ28 (Class IV; Bruggemann et al, 2006; Heuner, 2007; Albert-Weissenberger et al, 2010). The alarmone rapidly increases the level of fleQ, fleR and several Class II flagellar transcripts, including flgB (Table 1).
Kinetic analysis of flgB expression revealed the versatility of ppGpp and DksA control. In response to ppGpp, DksA is essential for flgB expression, but the amount and/or duration of the ppGpp stimulus governs the level of flgB RNA. As ppGpp accumulates, RNA for the Class II, σ54-dependent gene target flgB first accumulates and then decreases (Fig. 9A). In S. enterica, only six subunits of FlgB polymerize with other rod proteins prior to hook formation (Chevance & Hughes, 2008). Thus, the pulse of flgB RNA orchestrated by ppGpp and DksA may contribute to proper stoichiometry of rod and hook subunits.
In contrast to flgB, ppGpp and DksA continuously activate fliA (σ28) and flaA during differentiation (Fig. 9A), consistent with induction of fliA in the transmissive phase (Fig. 9B; Bruggemann et al, 2006). As alarmone increases (Hammer & Swanson, 1999), so do fliA and flaA RNA levels, suggesting functional FliA accumulates. FliA activates Class IV flagellar genes such as flaA and fliD, in addition to genes unrelated to flagellar biogenesis (Heuner et al, 2002; Molofsky et al, 2005; Bruggemann et al, 2006; Albert-Weissenberger et al, 2010). Continuous elevated activation of flaA by FliA is fitting, since the typical bacterial flagellum is comprised of ~ 20,000 filament subunits (Chevance & Hughes, 2008).
Phenotypic analyses also indicate that ppGpp modulates activation initiated by DksA. In response to fatty acid stress, DksA controls L. pneumophila flaA promoter activation and evasion of macrophage degradation independently of ppGpp (Fig. 7). When plasmid-borne dksA expression is induced, ppGpp0 bacteria initiate flagellar biosynthesis, motility and cytotoxicity to macrophages (Fig. 8). Furthermore, DksA mediates basal fliA promoter activity during E phase (Fig. 9B). Likewise, in both exponential and stationary phase E.coli, DksA regulates flagellar biogenesis directly, in this case by inhibiting transcription from a σ70-dependent fliA promoter (Lemke et al, 2009). Furthermore, when over-expressed, DksA suppresses several E. coli ppGpp0 mutant phenotypes, including amino acid auxotrophy, autoaggregation, motility and RpoS accumulation (Potrykus & Cashel, 2008). DksA can also regulate E. coli gene expression independently and even oppositely of ppGpp (Lyzen et al, 2009; Merrikh et al, 2009). Perhaps at the promoters of certain flagellar genes or their regulators (FleQ, σ54, FleR, and σ28), DksA controls RNAP activity independently of ppGpp.
It is noteworthy that E. coli and L. pneumophila each enlist DksA and ppGpp to regulate flagellar genes, but to the opposite effect. Commensal enteric E. coli utilize DksA and ppGpp to repress ribosome and flagella synthesis simultaneously (Lemke et al, 2009), whereas aquatic intracellular L. pneumophila have co-opted the stringent response to down-regulate ribosomal transcripts while activating flagellar genes, which promote transmission to a new replication niche. Judging from BLASTP analyses, L. pneumophila also lack the canonical chemotaxis components that equip E. coli to swim toward nutrients (data not shown). Because L. pneumophila is constrained within a host vacuole during replication, this pathogen presumably exploits whatever nutrients are available. Also, E. coli is equipped with numerous peritrichous flagella, whereas L. pneumophila synthesize a monopolar flagellum. Regardless of the distinct costs and benefits that motility confers to E. coli and L. pneumophila in their natural reservoirs, mechanistic studies can now exploit this dichotomy. Perhaps sequence differences observed in the N-terminal globular domain of the two DksA proteins (Fig. 2A) account for their distinct mode of regulation. Alternatively, the promoter architecture of key regulators like fliA may dictate the impact of each stress response.
To express certain transmission traits fully, L. pneumophila requires that DksA cooperate with ppGpp. For example, DksA only partially activates the flaA promoter and evasion of lysosomal degradation by ppGpp0 mutants in response to propionic acid (Fig. 7). The alarmone is also essential for activation of transmission phenotypes in response to acetate (data not shown) or entry into stationary phase (Figs. 3 and 7A). Moreover, dksA induction fails to rescue pigmentation (Fig. 6B), sodium sensitivity, infectivity and stationary phase survival defects of ppGpp0 mutants (data not shown). Whether ppGpp affects DksA protein levels has not been tested in L. pneumophila, but dksA RNA levels are similar during the replicative and transmissive phases in broth and amoebae (Bruggemann et al, 2006). The genetic studies presented here establish that the capacity of DksA and ppGpp to act independently or cooperatively during L. pneumophila differentiation is context-dependent.
Analysis of DksA and ppGpp also revealed that the requirements for L. pneumophila to transit from broth to macrophage and from macrophage-to-macrophage are distinct. Like DksA, the regulators LetA, LetS, LetE and FliA each activate transmission phenotypes in broth, yet are dispensable for transmission from one macrophage to another (Fig. 3; Hammer et al, 2002). In contrast, ppGpp-dependent factors are critical for L. pneumophila transmission from lysosomal vacuoles of permissive mouse macrophages (Sturgill-Koszycki & Swanson, 2000; Dalebroux et al, 2009). Perhaps in host cells, ppGpp induces L. pneumophila pathways that promote infection of A. castellanii (Fig. 4) and spread between macrophages (Fig. 5) but are poorly expressed in broth culture. In fact, of the cohort of genes upregulated during transmission in amoebae, only ~ 77% are also elevated during PE phase in broth (Bruggemann et al, 2006). It is also possible that L. pneumophila regulators control parallel, or perhaps even redundant pathways in particular host environments. Indeed, although dispensable for macrophage transmission, the FliA sigma factor and the LetA response regulator are required for bacterial growth in particular amoebae (Heuner et al, 2002; Gal Mor & Segal, 2003). Also, some bacteria use ppGpp to control cellular processes through physical interactions with factors other than RNAP (Wang et al, 2007; Zhao et al, 2008).
Synthesis of ppGpp by L. pneumophila rapidly alters the level of numerous transcripts. As in E. coli (Durfee et al, 2008; Traxler et al, 2008), ppGpp immediately down-regulates the protein synthesis machinery of L. pneumophila. Indeed, of the targets showing decreased RNA levels, nearly half encode ribosomal proteins and translation machinery (Table S1). While the impact of ppGpp on RNAP has been the focus of much research, the alarmone also induces growth arrest and may alter transcript stability, factors which will also affect the level of transcripts in a cell. Mechanistic studies of particular promoters and transcripts can now identify how ppGpp alters the L. pneumophila transcriptional profile during its life cycle.
In the transmissive state, L. pneumophila activates its Dot/Icm Type IV secretion system; this virulence mechanism is also coordinated in part by ppGpp. For example, genes encoding secreted substrates are activated in the PE phase (Nagai et al, 2002; Bruggemann et al, 2006; Tiaden et al, 2007; Rasis & Segal, 2009), and effectors such as LepA and LepB contribute to non-lytic release from amoebae (Chen et al, 2004). Consistent with these findings, RNAs for three secreted coiled-coil domain containing host cell effectors rapidly accumulate, including LepA (Table 1). Transcripts encoding membrane and channel components of the Dot/Icm system are also responsive to ppGpp. However, since many of these proteins are constitutively expressed during E and PE phase in broth (J. P. Vogel, personal communication), basal levels of ppGpp may mediate their expression, or more complex post-transcriptional mechanisms contribute.
L. pneumophila differentiation also entails a variety of metabolic changes. In the transmissive form, the pathogen activates several phospholipases, some of which are secreted into host cells via the Type II secretion (Lsp) and Type IV Dot/Icm secretion systems (Banerji et al, 2008). The level of a phospholipase (lpg2837) RNA immediately increases in response to ppGpp (Table 1). Additionally, transcripts for central metabolic and fatty acid activating enzymes like pta are up-regulated by ppGpp. Alarmone accumulation also leads to accumulation of an RNA encoding an uncharacterized type I polyketide synthase (Gokhale et al, 2007). During differentiation to the transmissive form, L. pneumophila modifies its surface and sheds vesicles that inhibit phagosome-lysosome fusion (Fernandez-Moreira et al, 2006). Concomitant with this process, several LPS modification and phospholipid biosynthesis enzymes are down-regulated by ppGpp (Table S1). These alterations in LPS structure and phospholipid content may be critical for transmission of progeny, since mutants that lack ppGpp are degraded at the end of the replication period (Fig 5B). Therefore, regulation of a number of lipid pathways by ppGpp is another component of L. pneumophila differentiation.
L. pneumophila also employs ppGpp and DksA to recruit other regulatory factors to orchestrate its cellular differentiation. In response to alarmone accumulation, L. pneumophila up-regulates transcripts for the stationary phase sigma factor RpoS (σ38) and the non-coding regulatory RNA RsmZ (Table 1). On the other hand, the amount of RsmY RNA did not increase in response to ppGpp. Therefore, our study supports the prediction that the RsmY and RsmZ regulatory RNAs exhibit varying degrees of sensitivity to ppGpp, possibly at the level of transcription initiation or RNA stability.
The L. pneumophila flagellar cascade illustrates how stringent response factors can cooperate to govern bacterial life cycles. The interplay between DksA and ppGpp is complex, as their roles are distinct at different loci. With the identification of candidate genes by microarray analysis, detailed kinetic studies in which ppGpp synthesis by WT and dksA bacteria is synchronized can reveal how ppGpp and DksA coordinate complex developmental processes.
EXPERIMENTAL PROCEDURES
Bacterial strains and culture
L. pneumophila strain Lp02 (thyA hsdR rpsL; MB110), a virulent thymine auxotroph derived from Philadelphia 1 (Berger & Isberg, 1993), was the parental strain for all the strains analyzed. L. pneumophila was cultured at 37°C with agitation in 5 ml of N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; Sigma)-buffered yeast extract (AYE) broth or on ACES-buffered charcoal yeast extract (CYE), supplemented with 100 µg/ml thymidine (AYET, CYET) when necessary. Bacteria from colonies <5 days old were cultured in broth overnight, then subcultured into fresh AYET prior to experiments. Exponential (E) cultures had an optical density at 600 nm (OD600) of 0.3 to 2.0; post-exponential (PE) cultures had an OD600 of 3.0 to 4.5, a period when the viability of the strains was similar (Fig. 2). Where indicated, ampicillin (amp; Fisher) was added to a final concentration of 100 µg ml−1; gentamycin (gent; Fisher) to 10 µg ml−1; chloramphenicol (cam; Roche) to 5 µg ml−1 for L. pneumophila and 10 ug ml−1 for E. coli; propionic acid to 10 mM; and, isopropyl-beta-D-thiogalactopyranoside (IPTG) to the concentrations specified. To determine colony forming units (CFU), serial dilutions of L. pneumophila were plated on CYET and incubated at 37°C for 4–5 days.
dksA mutant construction
To construct the dksA mutant, a deletion insertion allele was first generated by recombineering in E. coli (Datsenko & Wanner, 2000; Yu et al, 2000). pGEM-dksA was generated by amplifying the dksA region (lpg2338) using primers dksA1 and 2 (Table S2). The FRT::cat::FRT cassette was amplified from pKD3 using primers dksA-pKD3a and dksA-pKD3b. To generate pGEM-ΔdksA::FRT::cat::FRT, pGEM-dksA and the linear PCR product were co-electroporated into DY330, prepared as described (Yu et al, 2000), recombinants were selected on LB-cam, and the insertion verified by PCR. Plasmid DNA from DY330 recombinants was used to transform DH5α, and transformants were verified by PCR. Lp02 was transformed with the ΔdksA::FRT::cat::FRT allele by natural competence (Stone & Kwaik, 1999). Chromosomal recombination was confirmed by PCR, and the resulting ΔdksA::FRT::cat::FRT mutant L. pneumophila was designated MB699 (Table 3).
Table 3.
Bacterial strains and plasmids
| Strain | Relevant genotype/phenotype | Reference |
|---|---|---|
| E.coli | ||
| DH5α | F-endA1 hsdR17 (r- m+) supE44 thi-1 recA1 gyrA/ (Nalr) relA1 Δ(lacZYA-argF)U169Ф80dLacZΔM15λpirRK6 |
Laboratory collection |
| DY330 | W3110 ΔlacU169 gal490 λcl1857 Δ(cro-bioA) | (Yu et al, 2000) |
| L. pneumophila | ||
| MB110 | Lp02 wild-type, StrR, Thy-, HsdR- | (Berger & Isberg, 1993) |
| MB699 | Lp02 ΔdksA::FRT::cat::FRT mutant | This work |
| MB697 | Lp02 relA::gent spoT::kan double mutant | (Dalebroux et al, 2009) |
| MB698 | Lp02 p206-gent, vector control strain | This work |
| MB701 | Lp02 ΔdksA::FRT::cat::FRT mutant p206-gent | This work |
| MB731 | Lp02 ΔdksA::FRT::cat::FRT pdksAi-gent | This work |
| MB698 | Lp02 p206-cat, vector control strain | (Dalebroux et al, 2009) |
| MB686 | Lp02 relA::gent spoT::kan double mutant p206-cat | (Dalebroux et al, 2009) |
| MB687 | Lp02 relA::gent spoT::kan double mutant prelAL.p. | (Dalebroux et al, 2009) |
| MB700 | Lp02 relA::gent spoT::kan double mutant pdksAi-cat. | This work |
| MB359 | Lp02 prelAE.c. | (Hammer & Swanson, 1999) |
| MB702 | Lp02 ΔdksA::FRT::cat::FRT mutant prelAE.c. | This work |
| MB355 | Lp02 pflaAgfp td(Δ)I | (Hammer & Swanson, 1999) |
| MB732 | Lp02 ΔdksA::FRT::cat::FRT mutant pflaAgfp td(Δ)I | This work |
| MB685 | Lp02 relA::gent spoT::kan double mutant pflaAgfp td(Δ)I | (Dalebroux et al, 2009) |
| MB733 | Lp02 pfliAgfp td(Δ)I | This work |
| MB735 | Lp02 ΔdksA::FRT::cat::FRT mutant pfliAgfp td(Δ)I | This work |
| MB734 | Lp02 relA::gent spoT::kan double mutant pfliAgfp td(Δ)I | This work |
| Plasmids | ||
| pGEMT-Easy | MCS within coding region of B-lactamase α-fragment linearized with single-T overhanges, AmpR |
Promega |
| pGEM-Gent | pGEMT-Easy with 1.7 kb gentamycin cassette cloned into the MCS, source of 1.7 kb gentamycin resistance cassette, AmpR, GentR |
(Molofsky et al, 2005) |
| pKD3 | Template plasmid for λ Red system, bla FRT::cat::FRT, ori R6K | (Datsenko & Wanner, 2000) |
| pGEM-dksA | pGEMT-Easy with 1.4 kb PCR amplified dksA chromosomal region ligated into T overhangs, AmpR |
This work |
| pGEM-ΔdksA::FRT::cat::FRT | pGEM-dksA with 1.1 kb FRT::cat::FRT cassette from pKD3 recombineered using strain DY330 to delete and replace the dksA coding sequence leaving ~500 bp homology on either side for Lp02 chromosomal recombination, AmpR , CamR |
This work |
| pGEM-dksAi | pGEMT-Easy with 540 bp PCR amplified dksA chromosomal region ligated into T overhangs, AmpR |
This work |
| p206-cat | pMMB66EH derivative, Δmob lacIq,PtaclacUV5, CamR | (Morales et al, 1991) |
| pdksAi-cat | p206-cat with 540 bp SalI/HindIII fragment from pGEM-dksAi ligated between the SalI/HindIII sites in the MCS, colinear with the PtaclacUV5 promoter, lacIq, Inducible dksA expression, CamR. |
This work |
| p206-gent | p206-cat with the cat cassette replaced by the gentamycin resistance cassette from pGEM-Gent, GentR. |
This work |
| pdksAi-gent | p206-gent with 540 bp SalI/HindIII fragment from pGEM-dksAi ligated between the SalI/HindIII sites in the MCS, colinear with the PtaclacUV5 promoter, lacIq, Inducible dksA expression, GentR. |
This work |
| prelAE.c. | pMMB66EH derivative, lacIq,PtaclacUV5, GentR with a relA allele from E. coli encoding a truncated, metabolically active RelA, cloned into the MCS, colinear with the PtaclacUV5 promoter, lacIq Inducible relAE.c. expression, GentR. |
(Hammer & Swanson, 1999) |
| pflaAgfp | 150 bp flaA promoter fragment fused to GFPmut3 in pKB5 with Ptac and lacIq removed, td(Δ)i |
(Hammer & Swanson, 1999) |
| pfliAgfp | 304 bp fliA promoter fragment fused to GFPmut3 in pKB5 with Ptac and lacIq removed, td(Δ)i |
This work |
Inducible dksA expression
To generate strains in which expression of dksA could be induced, a promoterless fragment of dksA was cloned into either pMMB206-Δmob, a broad host range vector containing a PtaclacUV5 IPTG-inducible promoter with a cam cassette (p206-cat), or the same vector with a gent cassette (p206-gent). To construct pGEM-dksAi, the dksA locus was amplified from Lp02 using primers dksAi1 and i2. The fragment was excised from pGEM-dksAi and ligated into p206-cat or p206-gent immediately 3’ of the PtaclacUV5 promoter, generating pdksAi-cat and pdksAi-gent, respectively. Insertion was confirmed by PCR. For complementation experiments, MB699 transformed with pdksAi-gent were selected on gent, creating MB701 for inducible dksA expression (Table 3). To induce dksA expression by ppGpp0 mutant L. pneumophila, MB697 transformed with pdksAi-cat were selected on cam, creating MB700 for inducible dksA expression in the absence of ppGpp.
fliA-gfp promoter fusion
A fragment containing 304 bp 5’ of the fliA RBS (K. Heuner, personal communication) and encoding the putative σ70 promoter and transcriptional start was amplified using primers fliAP1 and P2. The fragment was ligated into pKB5 directly 5’of gfp as described (Hammer & Swanson, 1999). This plasmid was used to transform MB110, MB697 and MB699 (Table 3), generating MB733, MB734 and MB735, respectively.
Fluorometry
To monitor expression of the flagellin promoter, E phase cultures of MB355, MB685, and MB732 were diluted to OD600 (0.75–0.85) and treated with 10 mM propionic acid or water (t = 0; Fig. 7A). At the times indicated, the cell density of each culture was measured as OD600 and fluorescence was quantified as described (Edwards et al, 2009). To quantify fliA promoter activity, E phase cultures of MB733, MB734 and MB735 were diluted to OD600 = 1.0 (t = 0) and cultured to stationary phase. Fluorescence was detected as described above except that cultures were normalized to OD600 = 3.0.
Detection of ppGpp
Accumulation of ppGpp was detected by thin-layer chromatography (TLC) as described (Dalebroux et al, 2009). Briefly, E phase AYET broth cultures of ppGpp0 mutant bacteria carrying either pempty or prelAL.p were diluted to OD600 = 0.25 and labelled with ~ 100 µCi/ml of carrier-free [32P]-phosphoric acid at 37°C for 6 h (~ 2 generations). After labelling, the E phase cultures were treated with 500 µM IPTG and sampled at 30, 60, and 90 min post-induction.
Flagellin western analysis
After culture to OD600 = 1.0 in 50 ml, mutant bacteria were treated with 200 µM IPTG; untreated WT bacteria were the positive control. After culture to PE phase (~ 9 h indicated by * in Fig. 6A), cell pellets were harvested. Since induction of prelAE.c. immediately inhibited growth of dksA mutants, dksA pempty cultures were normalized to the dksA prelAE.c. culture density prior to pelleting. Bacteria were lysed with Qproteome™ kit (Qiagen), and equivalent volumes of ppGpp0 and dksA mutant L. pneumophila lysates were denatured and separated on a SDS-10% polyacrylamide gel. To avoid signal overload, WT cell lysates were diluted 1:100 in PBS prior to denaturing. Flagellin was detected using a 1:50 dilution of monoclonal antibody 2A5 (Molofsky et al, 2005).
Microarrays
To study the effect of ppGpp induction on L. pneumophila gene expression (Table 1), ppGpp0 mutants carrying either pempty, or prelAL.p., were grown in AYET to E phase OD600 = 1.4 before treatment with 500 µM IPTG. After 45 min (when the pempty and prelAL.p. cultures had increased to OD600 = 1.90 and 1.70 , respectively) and 90 min (OD600 = 2.45 and 2.10 for pempty and prelAL.p., respectively), cells were harvested, and total RNA was extracted as described (Milohanic et al, 2003). To test the contribution of DksA to PE phase expression of L. pneumophila flagellar genes (Table 2), WT and dksA mutant bacteria were normalized to an OD600 = 1.6 and cultured until 95–100% of the WT culture was motile. RNA was prepared from two independent cultures, and each RNA sample was hybridized twice with dye swap to the microarrays. The design of microarrays was based on all predicted genes of the genomes of L. pneumophila Paris, Lens and Philadelphia (Bruggemann et al, 2006) in addition to the ncRNAs rsmY and rsmZ (Sahr et al, 2009). Reverse transcription, labeling and hybridization were carried out as described (Sahr et al, 2009).
Data normalization and differential analysis were conducted as described (Sahr et al, 2009). If not stated otherwise, only differentially expressed genes with 2-fold changes were taken into consideration. Complete data sets are available at http://genoscript.pasteur.fr in a MIAME compliance public database maintained at the Institut Pasteur and were submitted to the ArrayExpress database maintained at http://www.ebi.ac.uk/microarray-as/ae/ under the Acc. No. pending.
Quantitative Real-time PCR
To validate the microarray data sets (Tables 1, 2 and S1), bacteria were cultured as described for Microarray analysis. To assess whether dksA contributes to ppGpp-dependent transcript accumulation (Fig. 9A), WT and dksA mutant bacteria carrying prelAE.c.were cultured to E phase OD600 (0.7–0.8) and treated with 500 µM IPTG. Bacteria were harvested at 0, 90 and 300 min post-IPTG. By 300 min, WT prelAE.c. bacteria had transformed to the transmissive state and were fully motile. To isolate RNA for Real-time PCR (RT-PCR), L. pneumophila cell pellets were lysed in TRIzol (Invitrogen), extracted with chloroform/isoamyl alchohol (24/1) and precipitated. Total nucleic acid concentration was assessed, and total nucleic acid was treated with Turbo DNA Free DNase (Ambion) to digest residual genomic DNA (gDNA). RNA integrity was assessed with an Agilent 2100 Bioanalyzer, and cDNA was synthesized using Superscript® II Reverse Transcriptase (RT) (Invitrogen) and random hexamers (New England Biolabs). To control for residual gDNA contamination, a no-RT cDNA synthesis reaction was run for each sample. Prior to qRT-PCR, cDNA was diluted to ~ 300 pg/ul (assuming a 100% RT yield) and ~ 1.5 ng of cDNA was added to the reaction mixture containing Brilliant II SYBR® Green Q-PCR Master Mix (Stratagene), a reference dye, and primers for each target at a final concentration of 300 nM, and reactions were run in triplicate. Non-template control reactions and dissociation curves were run for each primer pair. The lpg2096 locus served as an internal reference, as it showed no change in expression by microarray under any conditions or strains tested in these or other studies (data not shown). qRT-PCR was performed using an MX3000P instrument (Stratagene). For validation of the prelAE.c. (Table 1 and Table S1) and dksA/WT PE phase (Table 2) microarray data sets, letE, rpoS, flgB, fliA, flaA, fliD, ndk, lag-1 and lpg0260 (Table S2) were targeted (Figs. S1 and S2). To test the contribution of dksA to ppGpp dependent expression of flagellar genes (Fig. 9), flgB, fliA and flaA were targeted. All primers were designed to amplify 100–200 bp segments of cDNA.
Comparisons of the transcriptional profiles of PE phase WT and ppGpp0 mutants and of dksA and ppGpp0 mutants L. pneumophila were also sought. However, since these ppGpp0 mutant microarray data sets failed a series of independent qRT-PCR and promoter fusion validation tests, the data were not analyzed further.
Intracellular growth in A. castellanii
Three-day-old cultures of amoebae were washed in infection buffer (PYG 712 medium without tryptone, glucose, or yeast extract) and adjusted to 5 × 105 to 1.0 × 106 cells ml−1. PE phase AYE broth grown L. pneumophila harboring a plasmid conferring thymine prototrophy (pflaAgfp) were diluted in buffer and mixed with A. castellanii at an MOI of ~ 0.05. After allowing invasion for 1 h at 37°C, adherent amoebae were washed three times with infection buffer, and flasks were returned to 37°C. At the times shown, 300 µl aliquots from each flasks were centrifuged and vortexed to lyse intact amoebae, and CFU were enumerated by plating dilutions on CYE agar. Each infection was carried out in duplicate.
Macrophage cultures
Macrophages were isolated from femurs of female A/J mice (Jackson Laboratory) and cultured in RPMI-1640 containing 10% heat-inactivated fetal bovine serum (RPMI/FBS; Gibco BRL) as described (Swanson & Isberg, 1995). Following a 7-day incubation in L-cell supernatant-conditioned media, macrophages were plated at 5 × 104 per well for cytotoxicity assays or 2.5 × 105 per well for lysosomal degradation assays, infectivity assays and intracellular growth curves.
Infection and growth in macrophages
L. pneumophila binding, entry, and survival inside macrophages during a 2 h incubation was measured as described (Dalebroux et al, 2009). To complement the dksA mutant infectivity defect, plasmid carrying bacteria were induced with 25 µM IPTG in early E phase and cultured to PE phase prior to infection (Fig. 3A). To quantify intracellular growth, each pooled macrophage supernatant and lysate was plated for CFU at various times post-infection as described (Bachman & Swanson, 2001).
Degradation in macrophages
The percentage of intracellular L. pneumophila that remain intact after a 2 h infection was quantified by fluorescence microscopy (Bachman & Swanson, 2001). Except for longer incubation times (18 h) and MOI adjustments, identical procedures were used to image and score infected macrophages at later time points of the primary infection period (Fig. 5B).
Cytotoxicity
To measure contact-dependent cytotoxicity, L. pneumophila were added to macrophages at the indicated MOI, and cytotoxicity was measured spectrophoretically as described (Molofsky et al, 2005). To determine the contribution of dksA to PE phase activation of L. pneumophila cytotoxicity (Fig. 3C), WT pempty was cultured to E or PE phase, ppGpp0 pempty to PE phase, and dksA mutants from E to PE phase with 25 µM IPTG prior to infection. To determine if induction of dksA expression or constituitive ppGpp synthesis could restore ppGpp0 and dksA mutant cytotoxicity (Fig. 8), WT, ppGpp0, and dksA bacteria carrying pempty, pdksA, or prelAE.c. were cultured from E to PE phase (~ 9 h, indicated by * in Fig. 6A) in the presence of 200 µM IPTG prior to infection.
Sodium sensitivity
The percentage of L. pneumophila that were sensitive to sodium was determined by enumerating colony formation on CYET and CYET containing 100 mM NaCl as described (Byrne & Swanson, 1998).
Acknowledgements
We are grateful to Andrew Bryan for introducing us to recombineering, Dr. Paul Carlsen for qRT-PCR technical guidance, and Drs. David Friedman and Mary O’Riordan for sharing equipment. This research was supported by NIH grant 2 R01 AI44212 and a University of Michigan Rackham Predoctoral Fellowship.
REFERENCES
- Aberg A, Fernandez-Vazquez J, Cabrer-Panes JD, Sanchez A, Balsalobre C. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol. 2009;191(10):3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg A, Shingler V, Balsalobre C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol Microbiol. 2008;67(6):1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x. [DOI] [PubMed] [Google Scholar]
- Albert-Weissenberger C, Sahr T, Sismeiro O, Hacker J, Heuner K, Buchrieser C. Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J Bacteriol. 2010;192(2):446–455. doi: 10.1128/JB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alli OA, Gao LY, Pedersen LL, Zink S, Radulic M, Doric M, Abu Kwaik Y. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect Immun. 2000;68(11):6431–6440. doi: 10.1128/iai.68.11.6431-6440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Swanson MS. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol. 2001;40(5):1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- Bachman MA, Swanson MS. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect Immun. 2004;72(6):3284–3293. doi: 10.1128/IAI.72.6.3284-3293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay P, Byrne B, Chan Y, Swanson MS, Steinman HM. Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect Immun. 2003;71(8):4526–4535. doi: 10.1128/IAI.71.8.4526-4535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Aurass P, Flieger A. The manifold phospholipases A of Legionella pneumophila - identification, export, regulation, and their link to bacterial virulence. Int J Med Microbiol. 2008;298(3–4):169–181. doi: 10.1016/j.ijmm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J Bacteriol. 2009;191(2):616–624. doi: 10.1128/JB.01195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Bernardo LM, Johansson LU, Solera D, Skarfstad E, Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription. Mol Microbiol. 2006;60(3):749–764. doi: 10.1111/j.1365-2958.2006.05129.x. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Modulation of Chemical Composition and Other Paramters of the Cell by Growth Rate. In: Curtiss R III, Ingraham J, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2 ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006;8(8):1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- Byrne B, Swanson MS. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66(7):3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield CH, Cianciotto NP. The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect Immun. 2007;75(8):4062–4070. doi: 10.1128/IAI.00489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, de Felipe KS, Clarke M, Lu H, Anderson OR, Segal G, Shuman HA. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303(5662):1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6(6):455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto NP, Fields BS. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci U S A. 1992;89(11):5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol. 2008;67(3):619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE., 3rd The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003;100(17):10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Edwards RL, Swanson MS. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol. 2009;71(3):640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190(3):1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RL, Dalebroux ZD, Swanson MS. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol Microbiol. 2009;71(5):1190–1204. doi: 10.1111/j.1365-2958.2008.06593.x. [DOI] [PubMed] [Google Scholar]
- Faulkner G, Berk SG, Garduno E, Ortiz-Jimenez MA, Garduno RA. Passage through Tetrahymena tropicalis triggers a rapid morphological differentiation in Legionella pneumophila. J Bacteriol. 2008;190(23):7728–7738. doi: 10.1128/JB.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moreira E, Helbig JH, Swanson MS. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infect Immun. 2006;74(6):3285–3295. doi: 10.1128/IAI.01382-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O, Segal G. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb Pathog. 2003;34(4):187–194. doi: 10.1016/s0882-4010(03)00027-5. [DOI] [PubMed] [Google Scholar]
- Gokhale RS, Saxena P, Chopra T, Mohanty D. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat Prod Rep. 2007;24(2):267–277. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- Gummesson B, Magnusson LU, Lovmar M, Kvint K, Persson O, Ballesteros M, Farewell A, Nystrom T. Increased RNA polymerase availability directs resources towards growth at the expense of maintenance. EMBO J. 2009;28(15):2209–2219. doi: 10.1038/emboj.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Swanson MS. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33(4):721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Tateda ES, Swanson MS. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol. 2002;44(1):107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nature Rev Microbiol. 2008;6(7):507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) J Biol Chem. 1994;269(42):26584–26590. [PubMed] [Google Scholar]
- Heuner K, Albert-Weissenberger C. The flagellar regulon of Legionella pneumophila and the expression of virulence traits. In: Heuner K, Swanson M, editors. Legionella Molecular Microbiology. Norfolk, United Kingdom: Caister Academic Press; 2007. pp. 101–121. [Google Scholar]
- Heuner K, Dietrich C, Skriwan C, Steinert M, Hacker J. Influence of the alternative sigma(28) factor on virulence and flagellum expression of Legionella pneumophila. Infect Immun. 2002;70(3):1604–1608. doi: 10.1128/IAI.70.3.1604-1608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovel-Miner G, Pampou S, Faucher SP, Clarke M, Morozova I, Morozov P, Russo JJ, Shuman HA, Kalachikov S. SigmaS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol. 2009;191(8):2461–2473. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nature Rev Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74(6):1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Conover GM, Isberg RR. Legionella pneumophila EnhC is required for efficient replication in tumour necrosis factor alpha-stimulated macrophages. Cell Microbiol. 2008;10(9):1906–1923. doi: 10.1111/j.1462-5822.2008.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzen R, Kochanowska M, Wegrzyn G, Szalewska-Palasz A. Transcription from bacteriophage {lambda} pR promoter is regulated independently and antagonistically by DksA and ppGpp. Nucl Acid Res. 2009;37(20):6655–6664. doi: 10.1093/nar/gkp676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189(14):5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Ferrazzoli AE, Lovett ST. Growth phase and (p)ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli. J Bacteriol. 2009;191(24):7436–7446. doi: 10.1128/JB.00412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milohanic E, Glaser P, Coppee JY, Frangeul L, Vega Y, Vazquez-Boland JA, Kunst F, Cossart P, Buchrieser C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47(6):1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203(4):1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Shetron-Rama LM, Swanson MS. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect Immun. 2005;73(9):5720–5734. doi: 10.1128/IAI.73.9.5720-5734.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol. 2003;50(2):445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53(1):29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97(1):39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295(5555):679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61(1):194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118(3):297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Tedin K. The bacterial signal molecule, ppGpp, regulates Salmonella virulence gene expression. Mol Microbiol. 2004;52(6):1827–1844. doi: 10.1111/j.1365-2958.2004.04122.x. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Rasis M, Segal G. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol. 2009;72(4):995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06677.x. Published article March 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Bachman MA, Swanson MS. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc Natl Acad Sci U S A. 2005;102(28):9924–9929. doi: 10.1073/pnas.0502767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266(6):3760–3767. [PubMed] [Google Scholar]
- Sharma AK, Payne SM. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol Microbiol. 2006;62(2):469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138(1):146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BJ, Kwaik YA. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181(5):1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Swanson MS. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med. 2000;192(9):1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63(9):3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalewska-Palasz A, Johansson LU, Bernardo LM, Skarfstad E, Stec E, Brannstrom K, Shingler V. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on sigma54. J Biol Chem. 2007;282(25):18046–18056. doi: 10.1074/jbc.M610181200. [DOI] [PubMed] [Google Scholar]
- Thompson A, Rolfe MD, Lucchini S, Schwerk P, Hinton JC, Tedin K. The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J Biol Chem. 2006;281(40):30112–30121. doi: 10.1074/jbc.M605616200. [DOI] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Weber SS, Bruggemann H, Bosshard R, Buchrieser C, Hilbi H. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol. 2007;9(12):2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68(5):1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Roy C, Isberg RR. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann N Y Acad Sci. 1996;797:271–272. doi: 10.1111/j.1749-6632.1996.tb52975.x. [DOI] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128(5):865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97(11):5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Weatherspoon N, Kong W, Curtiss R, 3rd, Shi Y. A dual-signal regulatory circuit activates transcription of a set of divergent operons in Salmonella typhimurium. Proc Natl Acad Sci U S A. 2008;105(52):20924–20929. doi: 10.1073/pnas.0807071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman T, Gal-Mor O, Segal G. Characterization of a Legionella pneumophila relA insertion mutant and toles of RelA and RpoS in virulence gene expression. J Bacteriol. 2002;184(1):67–75. doi: 10.1128/JB.184.1.67-75.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]