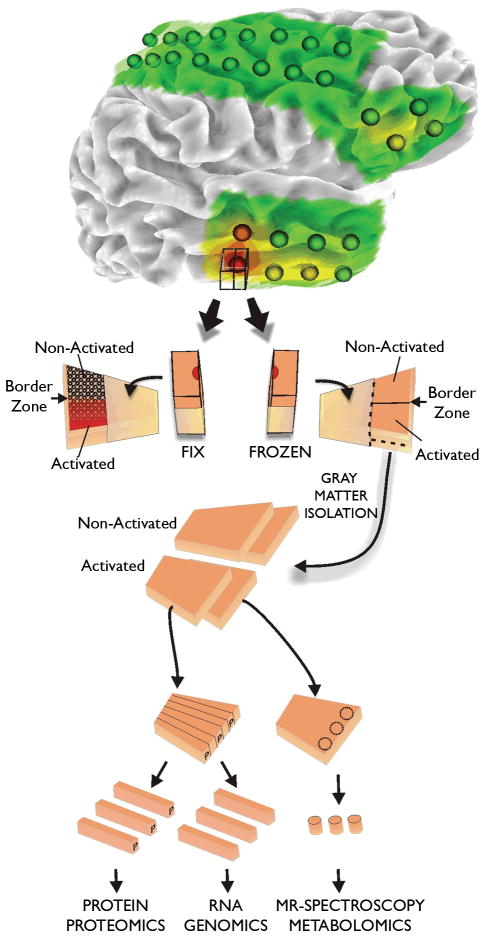
Figure 2. Tissue processing of human epileptic neocortical tissues.
All tissue samples are first identified on 3-dimensional surface maps made for each patients brain from MRI studies. Superimposed on each map are the recording electrode locations. In addition, color coded heat maps indicating the location of specific electrical parameters are placed on the rendering, such as interictal spike frequency shown here. Tissue samples underlying specific electrodes are first subdivided with half placed in paraformaldehyde for tissue sectioning and the other half stored frozen. This frozen piece is later subdivided into strips for preparation of RNA for genomics, protein for proteomics, and samples for metabolomics. In this way, many different systems can be analyzed from the same electrode location or from ‘activated’ subregions identified from the fixed tissues.