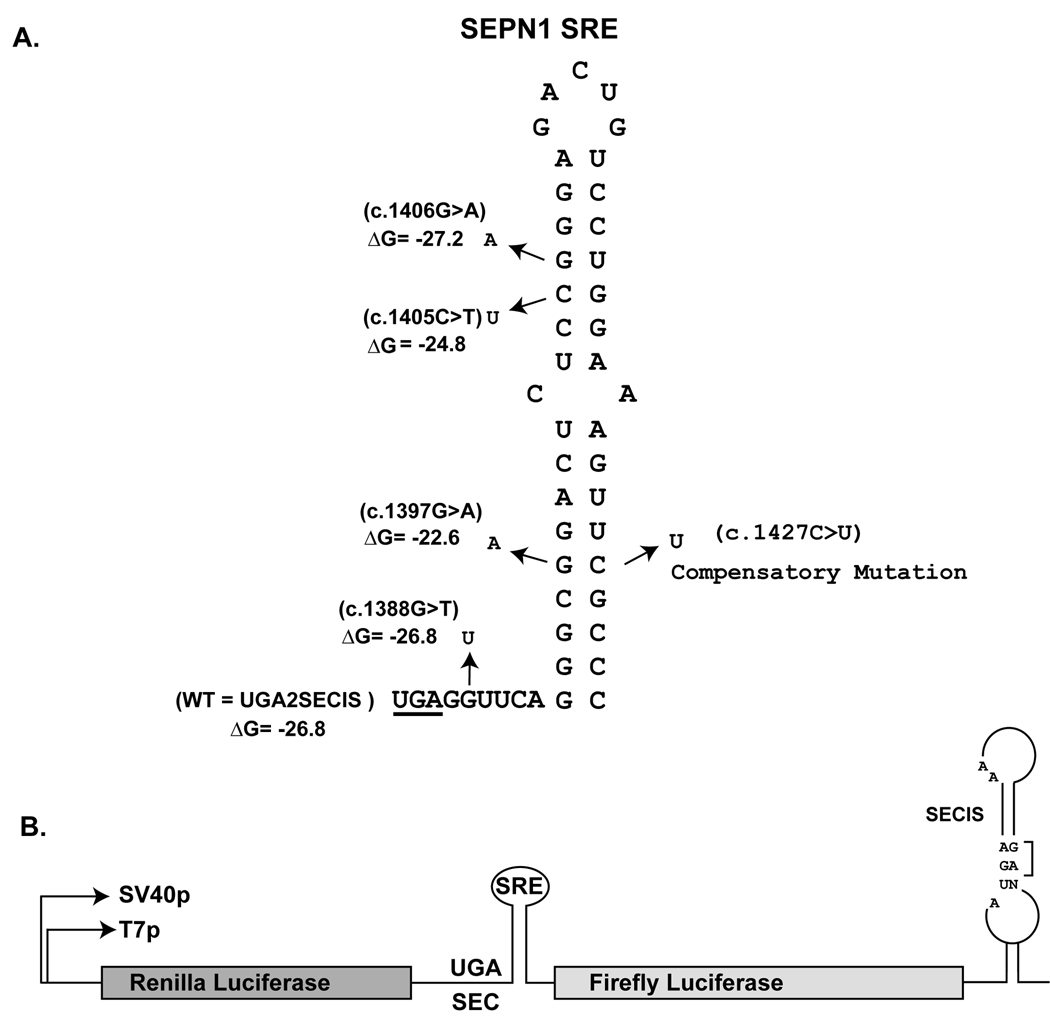
Fig. 1.
Stem-loop structure of the SEPN1 SRE. A) The UGA codon is underlined. The location and nucleotide identity of the four RSMD patient mutations are indicated by arrows. The predicted ∆G of the stem-loop is given in kcals/mole. The identity of the compensatory mutation introduced to restore base pairing between c.1397 and c.1427 is indicated. All numbering is based on cDNA sequence with the first nucleotide being the A of the ATG initiation codon. GenBank reference sequence for SEPN1 is NM_020451.2 B) Illustration of the Dual Luciferase (2luc) reporter vector. Renilla and Firefly coding sequences flank the SEPN1 SRE sequence and the SEPN1 SECIS is downstream of the Firefly open reading frame in the 3’UTR. UGA2SECIS referred to as WT in Figure 2 is the reporter vector with wildtype SEPN1 SRE and SECIS sequences. Essential features of the SECIS element such as the GA quartet and unpaired As in the loop are indicated.