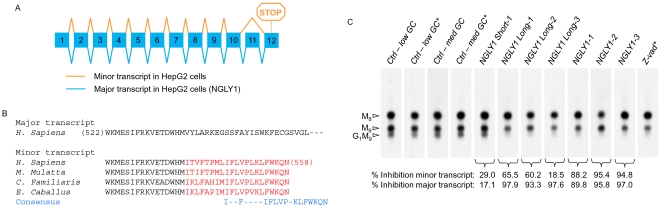
Figure 4. Alternative NGLY1 transcripts and the impact of their down regulation on fOS generation in HepG2 cells.
A. Genomic organization of the human NGLY1 gene and alternative transcripts found in HepG2 cells. The major transcript found in HepG2 cells is indicated in blue and corresponds to the already published NGLY1 mRNA sequence (NM_018297.3). A minor transcript indicated in orange is generated by skipping of exon 11 (NM_001145295.1). B. Partial alignment of homologous Ngly1p sequences (amino acids 522–558) encoded by the minor transcript and comparison with the corresponding region of the major NGLY1 transcript (accession numbers: Macaca mulatta; XP_001092914, Equus caballus; XP_001492093, Canis lupus familiaris; DN746692.1). Note that the isoform encoded by the minor transcript contains 21 amino acids in its COOH terminal which are not found in the peptide sequence encoded by the major transcript. C. HepG2 cells were transiently transfected with the different siRNA duplexes designed to silence either or both NGLY1 transcripts (see Table S1). Total RNA were extracted three days later and the mRNA levels of each specific transcript were analyzed by QPCR. In parallel, HepG2 cells were co transfected with ENG-3 and the different siRNA duplexes targeting the two NGLY1 transcripts. As described in Materials and Methods, because of the variable GC content of the different RNAi duplexes, two negative control siRNA duplexes with either medium (med GC) or low GC (low GC) content were used. After 3 days cells were pulse-radiolabeled with [2-3H]mannose for 30 min. In some experiments, cells transfected with ENG-3 alone or with control RNAi duplexes were treated with either Z-vad-fmk (40 µM) dissolved in DMSO (Z-vad*) or DMSO alone (*) for 45 min prior to, and during the radiolabeling period. Subsequently, [2-3H]fOS, [2-3H]LLO and [2-3H]Glycoproteins were extracted, purified and quantitated, and [2-3H]fOS were analysed by TLC. In order to take into account the differences in total incorporation of radiolabel into cells cultivated under the different conditions, a fraction of total fOS was loaded onto the TLC according to the ratio of the total cellular radioactivity for a given incubation to that recovered from the incubation incorporating least radioactivity. The scanned TLC lanes are from the same fluorograph, but due to uneven migration, the scans were aligned manually to facilitate interpretation of data. The migration positions of standard oligosaccharides are shown to the left of the chromatograms and the abbreviations used are as described in Fig 1. The percent inhibition of the major and minor NGLY1 transcripts provoked by the different RNAi duplexes was calculated using the QPCR data and is indicated underneath the appropriate lanes. This experiment was performed once.