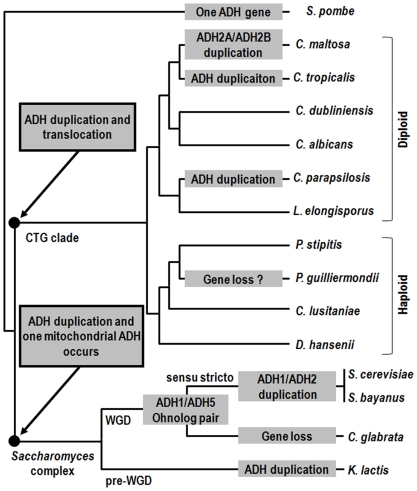
Figure 1. Minimum number of events required to explain evolution of ADH genes in some Saccharomycotina species.
ADH duplication events are shown in gray boxes. The topology of the phylogenetic relationships was a composite drawn from several sources [15], [46], [47]. Major clades were named, including the Saccharomyces complex, the CTG clade containing species that translate codon CTG as serine instead of leucine, the group of species that share the whole-genome duplication (WGD) and the Saccharomyces sensu stricto group. The ADH gene duplication and gene loss events in the CTG clade were deduced based on comparative analysis of the genomic contexts of ADH homologs from species of this clade (Figure S3). The ADH duplication events in the Saccharomyces complex were reported previously [6], [18], and confirmed with the genomic contexts of ADH homologs from the Yeast Gene Order Browser (YGOB), an online tool for visualizing comparative genomics of yeasts [47]. ADH1/ADH5 ortholog pair was retained in S. cerevisiae and one copy has been lost in Candida glabrata. K. lactis, a pre-WGD yeast, has duplicated the ADH genes independently more than once after separating from the post-WGD yeast species.