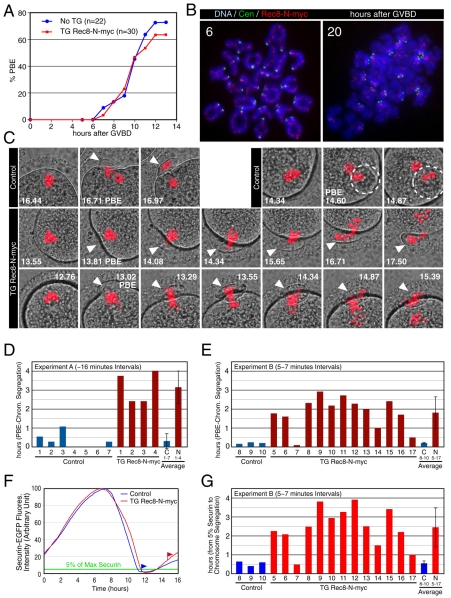
Fig. 9.
Rec8-N delays but does not block chiasma resolution at meiosis I in oocytes. (A) Kinetics of the first polar body extrusion (PBE) of oocytes cultured in vitro. The numbers of oocytes examined are indicated (n). (B) Localization of Rec8-N-Myc on chromosomes in TG Rec8-N-Myc oocytes. Chromosome spreads prepared from oocytes in culture for the indicated time after germinal vesicle breakdown (GVBD) were stained with anti-Myc antibody (red), CREST antiserum (green) and DAPI (blue). (C) Live confocal microscopy of oocytes expressing H2B-mCherry. DIC images (gray) were merged with images of the mCherry channel (red). Frames at indicated time (hours after beginning of filming) were selected from the original time series (supplementary material Movie 1). PBs are indicated by arrowheads or circles (when extruded vertically). (D) Intervals from PBE to the completion of chromosome segregation analyzed by ∼16-minute filming intervals (experiment A, supplementary material Movie 1). Error bar, s,d. (E) As in (D), analyzed by 5-7 minute intervals (experiment B, movies not shown). (F) Representative fluorescence quantifications of securin-EGFP signal in individual oocytes. The maximum value of the signal in each oocyte was set to 100 and relative intensity was plotted. Flags indicate time of the completion of chromosome segregation. Green line indicates the 5% level of the EGFP signal. (G) Intervals from 5% of the EGFP signal to the completion of chromosome segregation analyzed by 5-7 minute filming intervals (experiment B, movies not shown). Error bar, s.d.