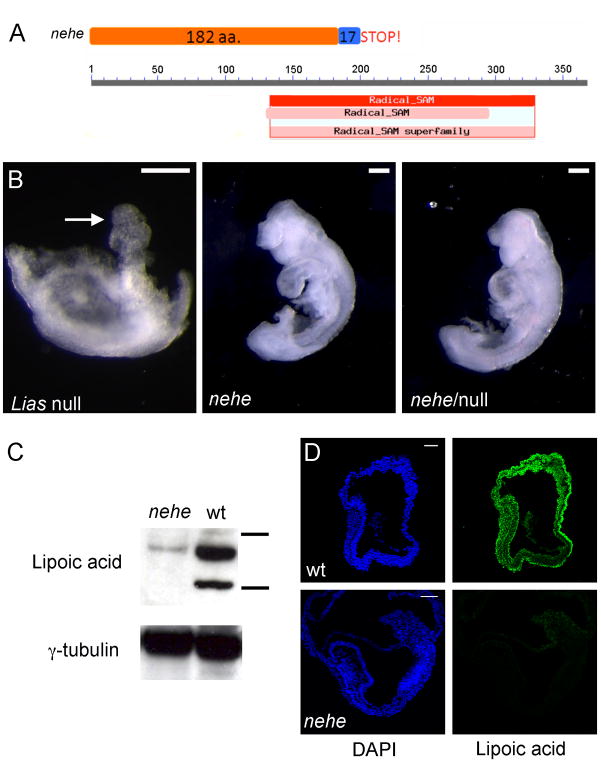
Figure 4. The nehe phenotype is caused by a hypomorphic mutation in Lipoic acid synthetase.
(A) The nehe mutation disrupts a splice site in Lias; translation of the mutant transcript would add 17 amino acids from the fifth intron to the open reading frame before a stop codon. The truncated protein made by the nehe allele lacks most of the radical SAM catalytic domain. (B) Liasnehe fails to complement the targeted null allele of Lias. Lias−/− embryos arrest at e7.5, with a poorly developed embryonic region and an allantois (arrow). In contrast, both Liasnehe homozygotes and Liasnehe/Liasnull embryos survive to later stages and show an anterior truncation and open anterior neural tube at e9.5. (C) Western blot analysis with an antibody that recognizes lipoic acid in e9.5 embryo extracts, compared to γ-tubulin levels. The horizontal bars next to wild-type lane represent 75 and 50 kD size markers. The two major substrates of lipoic acid are the E2 of KGDH, dihydrolipoamide succinyltransferase (Dlst), (predicted MW = 49 kDa) and the E2 of PGH, dihydrolipoamide S-acetyl transferase (Dlat) (predicted MW = 68 kDa). The levels of lipoic-acid modified proteins are greatly reduced in nehe embryos. (D) Immunofluorescent staining to detect Lipoic acid in e7.5 embryos shows that Lipoic acid-modified proteins are broadly distributed in wild-type embryos, but not detectable in nehe mutants.