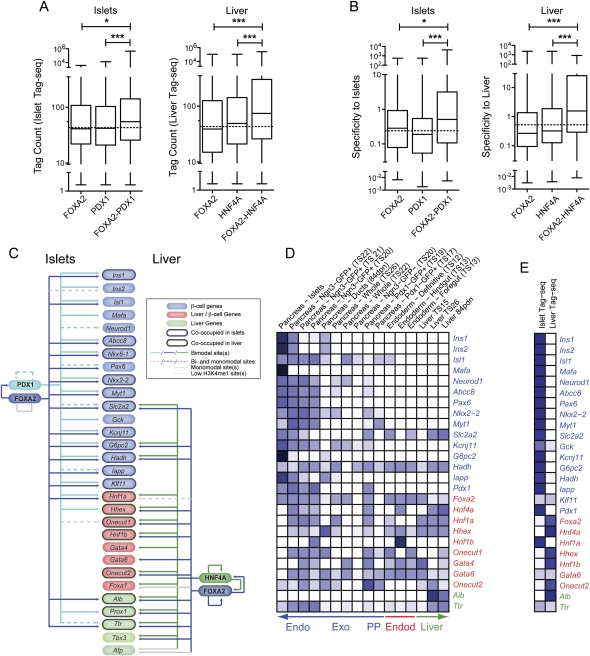
Figure 8.
Co-occupancy at bimodal sites drives tissue-specific gene expression. Box-whisker plots of islet or liver (A) Tag-seq counts or (B) specificity against 202 other mouse SAGE libraries, for expressed genes associated with bimodal sites either singly occupied by FOXA2, PDX1, or HNF4A, or co-occupied by FOXA2–PDX1 in islets, or FOXA2–HNF4A in liver. Statistically significant differences were detected using a Kruskall-Wallis non-parametric test with a Dunn's multiple comparison correction, with a P < 0.05 threshold. (*) P < 0.05; (***) P < 0.001. Bimodal FOXA2, PDX1, and/or HNF4A sites are commonly associated with islet and liver critical genes. (C) A schematic representing the presence of bimodal, monomodal, and low H3K4me1 FOXA2 (blue) and PDX1 (aqua) sites in islets (left), and of FOXA2 (blue) and HNF4A (green) sites in liver (right) associated with genes critical to islets (blue ovals), islets and liver (red ovals), or liver (green ovals) specification and/or function. The presence of one or more FOXA2, PDX1, or HNF4A bimodal sites associated with a gene is indicated by a solid colored line. The presence of one or more bimodal and one or more monomodal sites associated with a gene is indicated by a dotted colored line; the presence of monomodal sites only by a solid gray line, and of low H3K4me1 sites only by a dotted gray line. (D,E) Heatmaps of the relative expression levels (tag counts) of the indicated factors essential for proper pancreas and/or liver development or function in (D) SAGE libraries derived from endoderm, developing pancreas and liver, and adult pancreas and liver tissues, or (E) in the adult islet or liver Tag-seq libraries. Darker blue shades represent higher relative expression levels. Unambiguous tags above minimum count thresholds (four of the SAGE libraries and six for the Tag-seq libraries) for Foxa1, Nkx6-1, Prox1, Tbx3, and Afp could not be identified in the SAGE libraries.