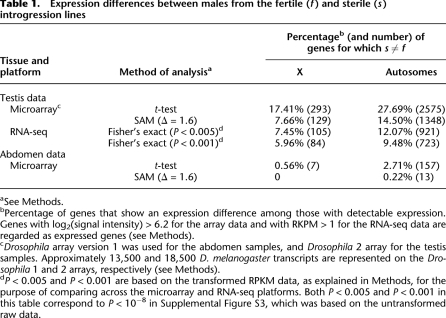
Table 1.
Expression differences between males from the fertile (f) and sterile (s) introgression lines
aSee Methods.
bPercentage of genes that show an expression difference among those with detectable expression. Genes with log2(signal intensity) > 6.2 for the array data and with RKPM > 1 for the RNA-seq data are regarded as expressed genes (see Methods).
cDrosophila array version 1 was used for the abdomen samples, and Drosophila 2 array for the testis samples. Approximately 13,500 and 18,500 D. melanogaster transcripts are represented on the Drosophila 1 and 2 arrays, respectively (see Methods).
dP < 0.005 and P < 0.001 are based on the transformed RPKM data, as explained in Methods, for the purpose of comparing across the microarray and RNA-seq platforms. Both P < 0.005 and P < 0.001 in this table correspond to P < 10−8 in Supplemental Figure S3, which was based on the untransformed raw data.