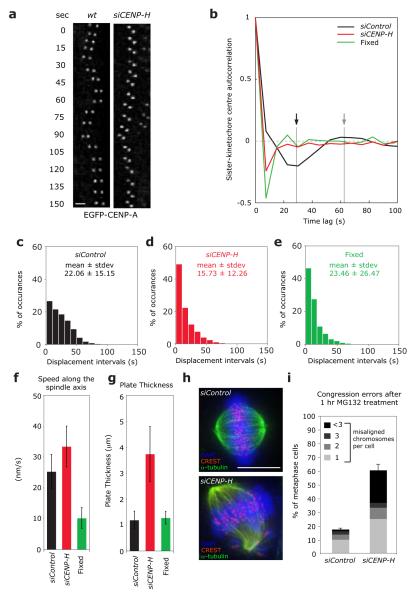
Figure 1.
Loss of CENP-H abolishes sister-kinetochore oscillations and disrupts metaphase plate alignment. (a) Live cell stills of single kinetochore pair oscillations in wild-type or CENP-H-depleted EGFP-CENP-A HeLa cells. Scale bar = 2 μm (b) Autocorrelation function of translational sister-kinetochore movements along the spindle axis (kinetochore oscillation) of siControl (black line), siCENP-H (red line) or fixed cells (green line). The autocorrelation function was calculated by combining all aligned sister-kinetochore pairs for each condition (Supplementary Information, Table S2). The first negative lobe in control-depleted cells (black arrow) indicates the half-period of the mean oscillation period, while its depth indicates the oscillation regularity. The second positive lobe (gray arrow) indicates the full period. Note that a random motion always produces a negative value at the first lag30 (c-e) Histograms of the mean interval time between directional switches of the sister-kinetochore pairs along the spindle axis of siControl (black bars), siCENP-H (red bars) or fixed cells (green bars). For each condition, the mean values and SDs are indicated. (f) Average sister-kinetochore pair speed along the spindle axis of siControl (black bar), siCENP-H (red bar) or fixed cells (green bar). Number of independent experiments n = 3. Error bars represent SD. (g) Width of the metaphase plate of siControl (black bar), siCENP-H (red bar) or fixed cells (green bar). Number of independent experiments n = 3. Error bars represent SD. (h) Representative images of control or CENP-H-depleted cells arrested in metaphase for 1 hour with the proteasome inhibitor MG132 and stained with DAPI (DNA; blue), α-tubulin (MTs; green) and CREST antisera (kinetochores; red). (i) Percentage of cells with one, two, three, or more than three uncongressed chromosomes following a 1 hour treatment with MG132 calculated from images such as shown in (h). Chromosomes in metaphase cells were counted as unaligned if they were located outside the central 30% of the mitotic spindle. Number of independent experiments n = 3 with 20 cells each. Scale bar = 10 μm. Error bars represent SEM.