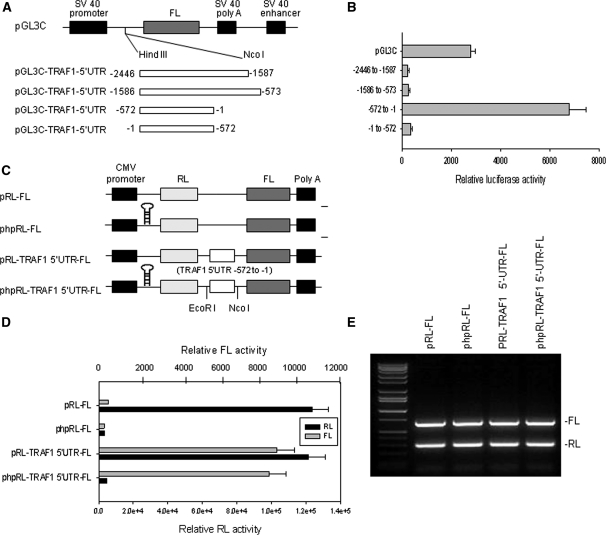
Figure 2.
Analysis of IRES activity in the 5′-UTR of TRAF1. (A) Schematic representation of the monocistronic plasmids used for the reporter assays. The entire 2446 bases TRAF1 5′-UTR was divided into three sections (−2446 to −1587, −1586 to −573 and −572 to −1) and each was cloned into pGL3C plasmid immediately upstream from the translation start codon of the firefly luciferase (FL) gene. (B) Transfection and reporter assay for IRES activity of TRAF1 5′-UTR. Raji cells were each co-transfected with 5 μg of pGL3C-TRAF1-5′UTR plasmid and 1 μg of pRL-SV40 vector as an internal control, by electroporation, and then quantitative RL and FL activities were detected using the Dual-Luciferase Reporter System. Data represent the mean ± SD of three independent experiments normalized to RL activity. (C) Schematic representation of the dicistronic constructs used for the reporter assays. The phpRL-FL is similar to pRL-FL, but with the addition of a stable hairpin at the 5′-end of the RL transcript to suppress cap-dependent translation of RL. (D) RL and FL activity analyses following transfection of Raji cells with 5 μg of each plasmid indicated, including pRL-FL and phpRL-FL vectors and constructions of these vectors with insertion of the fragment −572 to −1 of the TRAF1 5′-UTR. (E) RL and FL mRNA levels analyses following transfection of cells with various plasmids as described in (D). Total RNA was extracted, and RT–PCR was performed with specific primers targeted to FL and RL.