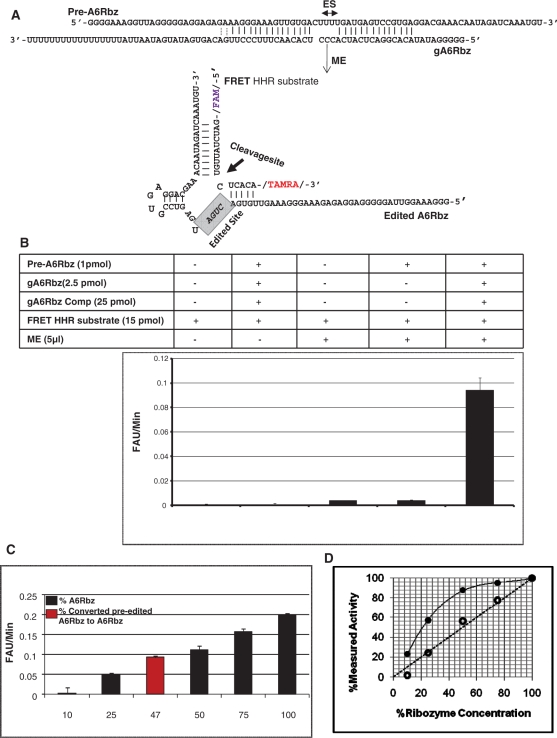
Figure 3.
FRET-based RNA editing assay. (A) Diagram of the pre-edited ribozyme (pre-A6RZ) is shown in association with the gA6RZ gRNA that specifies the deletion of three Us from the editing site (ES). In the presence of functional mitochondrial extract (ME), the pre-A6RZ is edited to active A6RZ that can now cleave the FRET HHR substrate with a fluorescent reporter (FAM) on the 5′-end and a quencher (TAMRA) on the 3′-end. The cleavage site of FRET HHR substrate is indicated by an arrow. (B) FRET-based RNA editing assay. Reactions were performed in a 30µl reaction volume under multiple turn-over conditions. Rotor-Gene 3000TM (CORBETT research) was used as a fluorescent reader. In the graph, y-axis represents the initial velocity of reaction in terms of fluorescent arbitrary unit (FAU) per minute and x-axis indicates the reaction conditions. A high signal-to-noise ratio was measured when both gA6RZ and ME were present (compare complete reaction with other conditions). All reactions containing gA6Rbz competitor (gA6Rbz Comp) were at 10× molar excess to gA6Rbz. The error bars represent the experimental variation (standard deviation) from 20 repetitions and resulting in Z-value of 0.65. (C) Various concentrations of active HHR, A6Rbz, 0.1, 0.25, 0.5, 0.75 and 1 pmol corresponding to 10%, 25%, 50%, 75% and 100% RNA editing was compared to efficiency of edited pre-A6Rbz FRET-based RNA editing assay. The red column indicates that nearly 47% of inactive HHR was edited to active HHR in the presence of gA6Rbz gRNA and ME. (D) FRET-based assay provides a linear measure of ribozyme activity. Close circles represent the measured ribozyme activity based on radiolabeled assay, whereas open circles indicate the measured ribozyme activity based on FRET assay. The dotted line represents a perfectly theoretical linear assay.