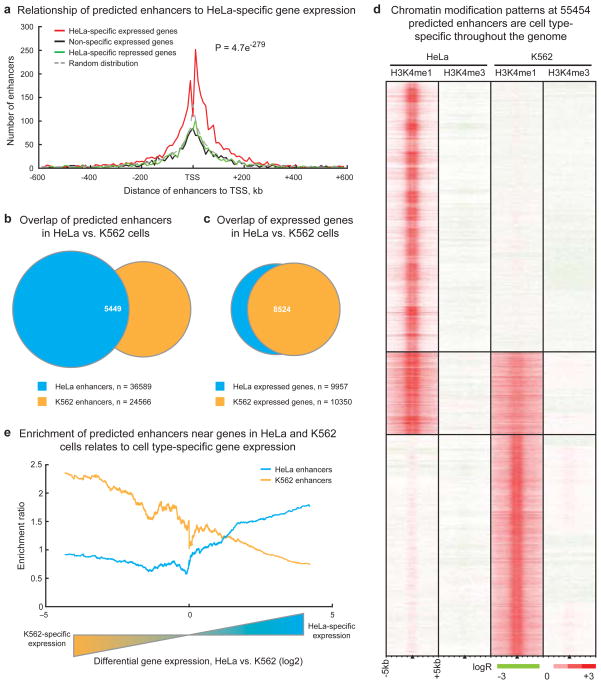
Figure 3. Chromatin modifications at enhancers are globally related to cell type-specific gene expression.
(A) Enhancer localization relative to genes that are HeLa-specific expressed compared to K562, GM06990, and IMR90 cells (red), non-specific expressed (green), HeLa-specific repressed (black), and a random distribution (dashed grey). Predicted enhancers are enriched around HeLa-specific expressed genes within insulator-defined domains and depleted in domains of ubiquitous or non-expressed genes (p-value reflects significance of enhancer enrichment in domains of HeLa-specific expressed genes, see Supplementary Information). (B) Most enhancers predicted in HeLa and K562 cells are cell-type specific while (C) most genes in HeLa and K562 cells are not specifically expressed; n = integer number of enhancers or genes in each set. (D) Chromatin modification patterns are cell type-specific at the majority of 55454 enhancers predicted in HeLa and K562 cells. (E) Comparison of enhancer enrichment and differential gene expression between HeLa cells and K562 cells revealed that HeLa enhancers are enriched near HeLa-specific expressed genes (blue line) while K562 enhancers are enriched near K562-specific expressed genes (orange line).