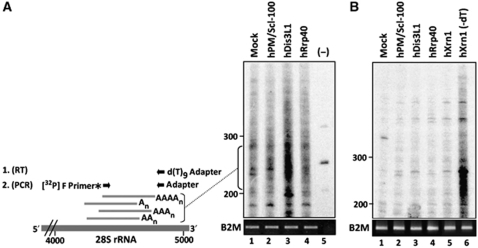
Figure 5.
Cytoplasmic accumulation of adenylated degradation intermediates after hDis3L1 knockdown. (A) Cytoplasmic RNA from cells in which the indicated proteins were silenced by RNAi was subjected to oligo(dT) RT–PCR labelling, followed by fractionation by denaturing polyacrylamide gel electrophoresis and autoradiography. A schematic representation of the labelling procedure is shown on the left. First, cDNA was generated using a d(T)9-adapter primer. Products from these reactions were amplified by PCR using a [32P]-labelled forward (F) primer corresponding to a sequence element of the 28S rRNA and a reverse primer corresponding to the adapter sequence. Material from a control reaction, in which the reverse transcriptase was omitted (−), is shown in lane 5. The positions of dsDNA markers are indicated on the left of the gel. For normalization, RT–PCR products obtained with PCR primers specific for the B2M gene are shown in the lower panel. (B) A similar oligo(dT) RT–PCR-labelling experiment as in panel A, but now the isolated RNA was incubated with oligo(dT) and RNase H before the RT reactions. RNA isolated from cells in which hXrn1 was knocked down, which similar to silencing of hDis3L1 leads to accumulation of poly(A) containing degradation intermediates, was used as a control to show that treatment with RNase H in the absence of oligo(dT) (lane 6) did not lead to the disappearance of the intermediates.