Fig. 1.

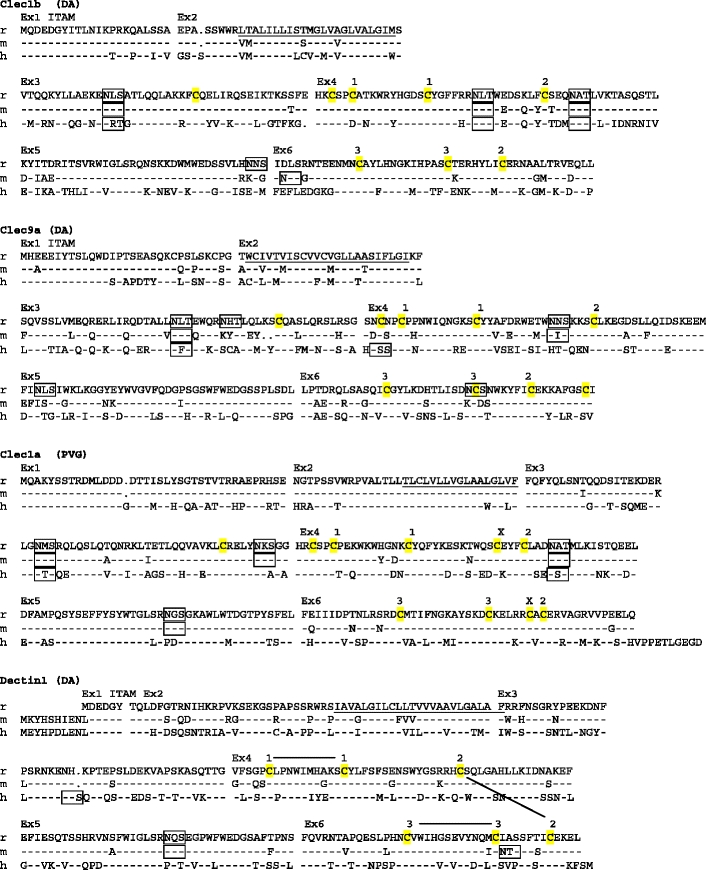
Amino acid sequence alignments of the CLSF receptors. r rat, m mouse, h human. Dashes mean identity with top sequence. Points indicate gaps. Ex1, Ex2, etc. on top of alignment show start site of corresponding exons. Predicted transmembrane regions (TMpred) are underlined. ITAMs and ITIMs are indicated. Potential N-glycosylation sites are boxed. Membrane external cysteines are highlighted in yellow. Numbers connected with black lines on top of the CD69 and Dectin1 sequences indicate SS-bonds as demonstrated for crystal structures of human CD69 and mouse Dectin1 (http://www.ncbi.nlm.nih.gov/ Genbank/). For the other genes, the corresponding cysteines are similarly numbered. X above cysteines in Clec1a indicates probable extra disulphide bridge, which in addition to the bridge between the cysteines labeled 2, links the first α helix with the last β strand of the lectin-like domain. GenBank accession numbers: rat CD69–GU357488, rat Clec2m–GU357487, mouse Clec2m–GU553093, rat Clec12a–GU357484, rat Clec12b–GU357483, rat Clec1b–GU357482, rat Clec9a–GU357486, rat Clec1a–GU357481, rat Dectin-1–GU357485. The rat strains from which the sequence was derived are shown in parentheses (PVG, DA, or both). It should be noted that the reported coding sequences were identical to the corresponding parts of the genomic rat sequence based on the BN strain. The single exception was rat Dectin1, with two single base substitutions in exon 6, one silent and one missense giving rise to an A225G substitution (identical for four independent cDNAs sequenced)
