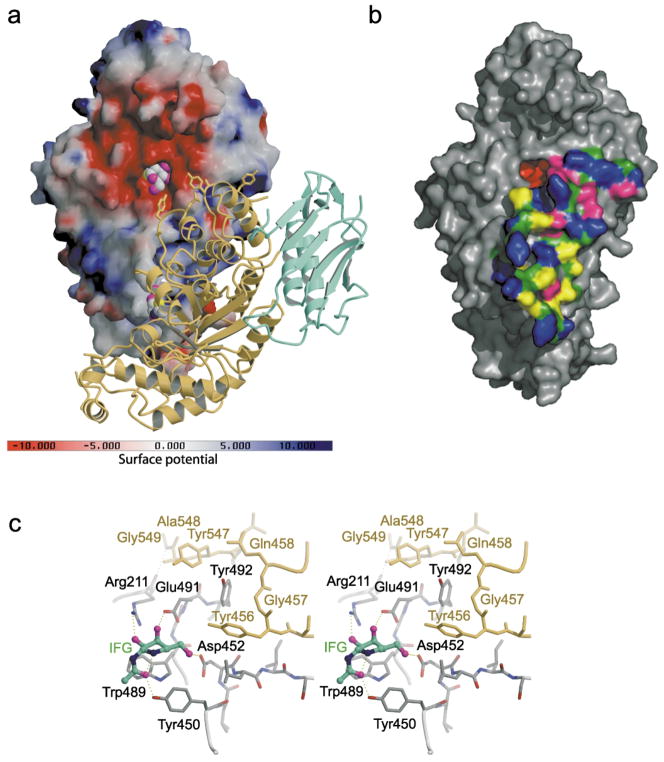
Figure 3.
Electrostatic potential surface map and dimer interface of human Hex B. (a) A solvent-accessible surface, drawn over one β-subunit and colored with regions of positive charge in blue and negative charge in red, reveals an overall negative charge about the active site. Note, however, that the electrostatic surface was calculated using the following charge profile only: Lys atom Nζ (charge 1.0); Arg atom NH1 (charge 0.5) and NH2 (charge 0.5); Glu atom Oε1 (charge −0.5) and Oε2 (charge −0.5); Asp atom Oδ1 (charge −0.5) and Oδ1 (charge −0.5); His atom Nδ1 (charge 0.5) and Nε2 (charge 0.5); OXT (charge −1.0). Due to the acidic environment of the lysosome, the electrostatic surface potential of Hex B in the lysosome may be slightly different from what is represented here, potentially being less negatively charged about the active site due to protonation of Glu and Asp carboxyl groups (surface created using the program GRASP84). The other subunit of the homodimer is represented by a ribbon diagram with domain I in green and the catalytic (β/α)8 domain II in yellow. The intermediate analogue NAG-thiazoline, bound in the active site of each subunit is shown as a space-filling model with carbon atoms in gray, oxygen in magenta, nitrogen in blue and sulfur in yellow. (b) Surface rendering of a single β-subunit showing the extensive surface area buried at the dimer interface as determined using the CNS program.74 Polar side-chains are colored blue, hydrophobic side-chains in yellow, backbone atoms in forest green, charged residues in magenta and residues not involved in dimerization are colored gray. The active site pocket is colored red ((b) was drawn using the program PyMOL85). (c) Active site residues (gray) stabilized by interactions from residues of the partnering subunit (yellow). The 2-fold symmetry at the dimer interface results in both active sites experiencing the same stabilizing effects from the associated monomer. The crystallographically determined position of GalNAc–isofagomine (IFG) in the active site of each subunit demonstrates that four of the six hydrogen bonds between the enzyme and inhibitor depend on stabilizing interactions from the partnering subunit. In the absence of the protein–protein interactions that are formed upon dimerization, Arg211, Glu491, Asp452 and Tyr450 are most likely too unstructured to be catalytically acitve.