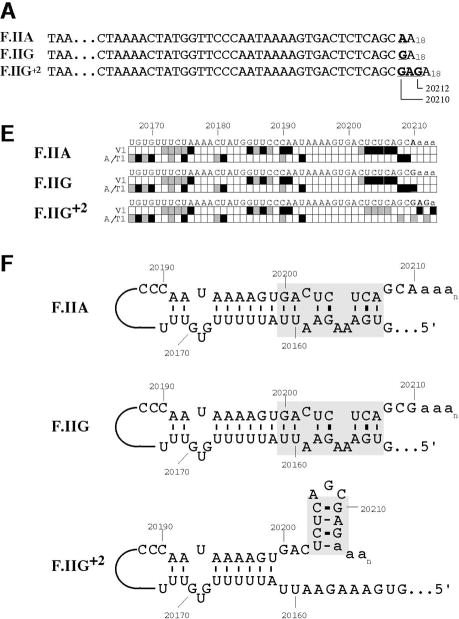
Fig 3. Secondary structures of F.II 3′UTRs with different 3′-processing sites.
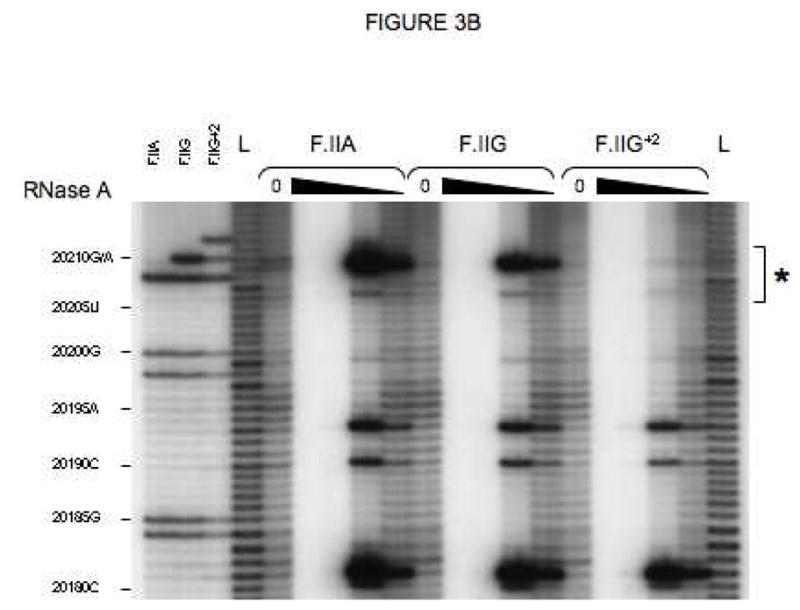
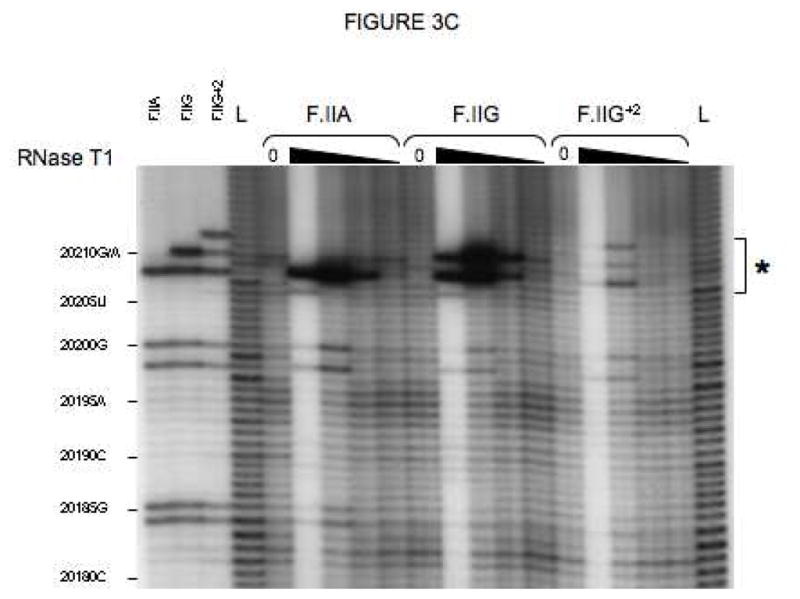
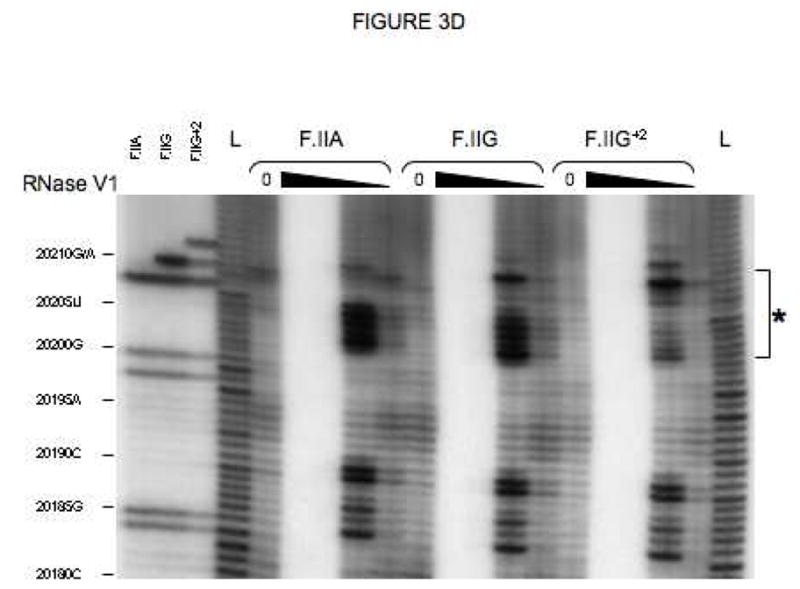
(A) Probes used for enzymatic structure-mapping studies. [32P]-labeled RNAs corresponding to three representative F.II 3′UTRs (F.IIA, F.IIG, F.IIG+2) were transcribed in vitro from the cognate DNA templates. All three RNAs contain identical polyadenylate (A)18 tails. Primary sequence differences are bolded and underlined. (B–D) Enzymatic secondary-structure mapping of F.II 3′UTRs. 5′-end [32P]-labeled F.II 3′UTRs were digested with RNase A (panel B), RNase T1 (panel C), and RNase V1 (panel D) at each of four different concentrations (wedges). Reaction products were resolved on an acrylamide-urea gel, and exposed to film. Nucleotide assignments (left) were deduced from comparison to denatured, RNase T1-treated 3′UTRs (F.IIA, F.IIG, F.IIG+2) and to an alkaline hydrolysis ladder (L). Regions exhibiting differential sensitivities to individual RNases are indicated with an asterisk. (E) 20210- and 20212-terminal F.II 3′UTRs exhibit different secondary structures. The 3′-terminal sequences of the F.IIA, F.IIG, and F.IIG+2 3′UTRs are illustrated (uppercase), along with the initial portion of the poly(A) tail (lowercase). Nucleotides exhibiting high and intermediate RNase sensitivities are indicated by black and grey boxes, respectively. (F) In silico structural mapping studies. Computational analyses were refined by constraining defined nucleotides to single- or double-stranded interactions based upon the experimental results from panels B and C. Relevant features of the predicted F.IIG and F.IIG+2 structures are illustrated. Elements of stem structures encompassing the terminal F.II 3′UTR are boxed in gray.