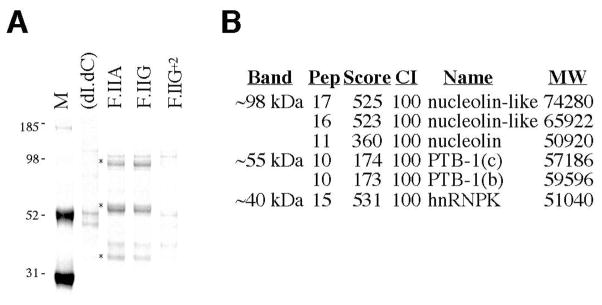
Fig 4. Differential binding of cytoplasmic factors to 20210- and 20212-terminal F.II 3′UTRs.
(A) Chromatographic analysis of HepG2 extract. Molar equivalents of streptavidin-ligated ssDNAs corresponding to F.IIG, F.IIA, and F.IIG+2 3′UTRs were incubated with HepG2 cell extract, and adherent proteins resolved by SDS-PAGE. Proteins that differentially bind 20210- and 20212-terminal F.II 3′UTR probes are indicated (asterisks). Agarose-liganded poly(dI.dC) ssDNA was assessed as a specificity control. M=protein size standards (kDa). (B) Mass spectrometric analysis of 3′ cleavage site-dependent binding factors. Tryptic digests of gel slices containing the three bands of interest were resolved by TOF-TOF analysis. For each identity, the peptide species count (Pep), protein score (Score), and confidence of identification (CI) are indicated. Score and CI values that are >70 and >95, respectively, are considered highly significant; other identities not meeting either criteria have been excluded from the figure to preserve clarity. MW=calculated molecular weight (Da).