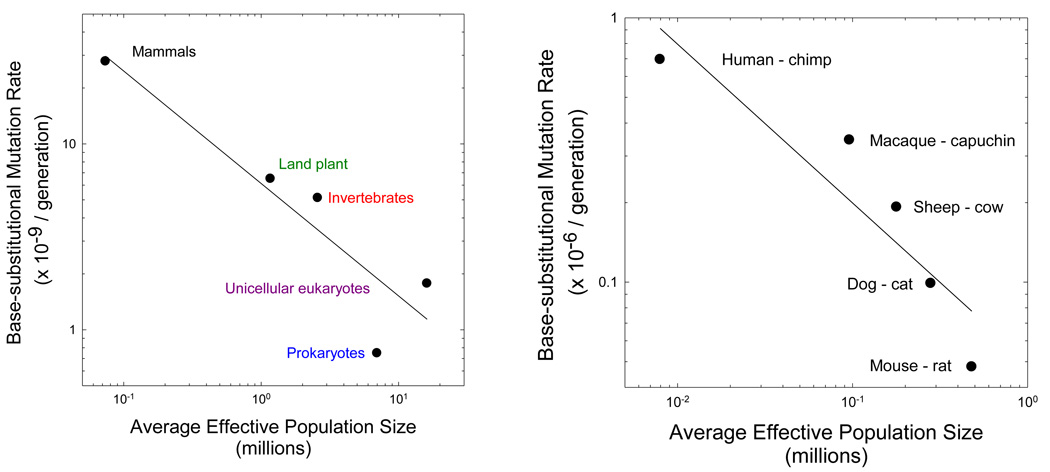
Figure 2.
The scaling of the base substitution mutation rate per generation (u) and the effective number of genes per locus (2Ne for diploids, and Ne for haploids). (a) The slope of the log-log regression for the nuclear genome of major phylogenetic groupings is −0.60 (0.16), where the number in parentheses denotes the standard error, with r2 = 0.84, although if the estimated Ne for prokaryotes is assumed to be 10 times too low (Lynch 2007), the slope is modified to −0.52 (0.02) with r2 = 0.99. (b) The slope of the log-log regression for the mitochondrial genome of mammalian lineages is −0.60 (0.15), with r2 = 0.84. The data are the average estimates from analyses assuming fixed and variable substitution rates in Piganeau and Eyre-Walker (2009).