Figure 5.
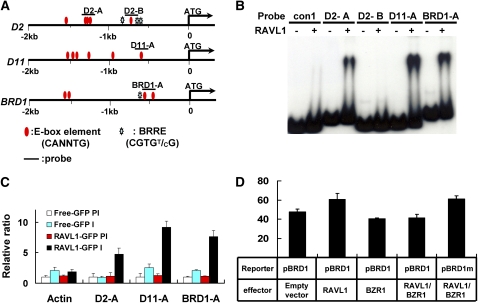
In Vitro and in Vivo Assay of RAVL1 Affinity to E-Boxes of the Promoters of BR Synthetic Genes (D2, D11, and BRD1).
(A) The schematic indicates the locations of the E-box and BRRE candidate elements and of the probes used in the EMSA and ChIP assays.
(B) EMSA for BR synthetic gene (D2, D11, and BRD1) promoters was performed using the D2-A, D2-B, D11-A, and BRD1-A probes, as shown in the schematic. For comparison with the data for BRI1 (Figure 4A), the con1 probe is the same as that used in EMSA with the BRI1 promoter.
(C) For ChIP assay, DNA immunoprecipitated using a GFP antiserum was amplified to detect E-box regions in the promoters of BR synthetic genes. The relative ratios of immunoprecipitated DNA to input DNA were determined by qRT-PCR. Input DNA was used to normalize the data. Error bars are se of the means of three qPCR replicates.
(D) The activity of native and BRRE-mutated promoters with different combinations of RAVL1 and BZR1. A luciferase gene driven by a 35S promoter is an internal control that was used to normalize GUS expression data. Error bars are se of the means of five replicates. BRD1 is the native promoter, and BRD1m contains a mutation of the BRRE element (−669CGTGTG−664) to CCCGGG.
[See online article for color version of this figure.]