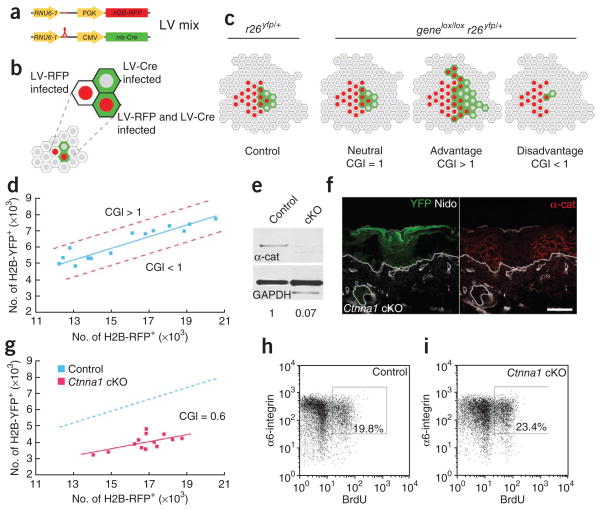
Figure 3.
Rapid assay for measuring an epidermal growth advantage or disadvantage conferred by a gene mutation reveals an unexpected growth disadvantage following α1-catenin loss despite hyperproliferation. (a–c) Schematic of the cellular growth index (CGI) assay. E9.5 Cre reporter embryos are infected with a mix of LV-Cre and LV-RFP, resulting in epidermal cells that have been transduced and that express H2B-RFP, H2B-YFP or both (a,b). At E18.5, the relative ratios of H2B-RFP+ to YFP+ cells in control (r26yfp/+) and gene knockout (genelox/loxr26yfp/+) mice are compared (c). Phenotypes are scored as either being neutral or having a growth advantage or disadvantage depending on this CGI value. (d) Graph of FACS-quantified numbers of H2B-RFP+ cells relative to YFP+ cells in control mice at E18.5. Genes whose depletion results in a growth advantage or disadvantage would shift the curve toward the upper or lower dashed red lines, respectively. (e) Quantified anti–α1-catenin (α-cat; test) and glyceraldehyde phosphate dehydrogenase (GAPDH; control) immunoblots of protein lysates from cells FACS-sorted from LV-Cre–infected control and Ctnna1-floxed (cKO) embryos. (f) Back skin sections of LV-Cre Ctnna1lox/lox r26yfp/+ (Ctnna1 cKO) embryos immunolabeled with anti–α1-catenin. Transduced cells are indicated by their YFP expression. (g) Graph of numbers of H2B-RFP+ cells relative to YFP+ cells in control (as in d) and Ctnna1 cKO mice at E18.5. Note the reduced CGI (0.6; P < 0.001) in the Ctnna1 cKO clones. (h,i) FACS plots and quantification of % basal (α6-integrin+) cells that incorporated BrdU after a 6-h labeling of E18.5 embryos. Note elevated BrdU incorporation despite the growth disadvantage in Ctnna1 cKO skin. Nidogen (Nido) marks the epidermal-dermal boundary as well as dermal blood vessels. Scale bar, 50 μm.