Abstract
We envisioned that label-free control of the transport of cells in two dimensions through receptor-ligand interactions would enable simple separation systems that are easy to implement, yet retain the specificity of receptor-ligand interactions. Here we demonstrate nanomechanical control of cell transport in two dimensions via transient receptor-ligand adhesive bonds by patterning of receptors that direct cell rolling through an edge effect. HL-60 cells rolling on P-selectin receptor patterns were deflected at angles of 5 – 10° with respect to their direction of travel. Absence of this effect in the case of rigid microsphere models of cell rolling suggests that this two-dimensional motion depends on nanomechanical properties of the rolling cell. This work suggests the feasibility of simple continuous-flow microfluidic cell separation systems that minimize processing steps and yet retain the specificity of receptor-ligand interactions.
Techniques for separation of cells rely on the ability to control their transport based on specific characteristics such as size, density, or surface ligands. In particular, control of transport in two dimensions is highly advantageous for the design of continuous-flow separation systems1–3. Continuous-flow separation of cells based on specific receptor-ligand interactions is currently limited to external control techniques that harness dielectrophoretic, magnetic, or other forces4, 5 and typically rely on capturing or labeling of the cells. These label-based approaches are very useful for cell separation, but involve special apparatus and multiple processing steps for labeling and label removal which are not desirable for sensitive cell samples or for point of care diagnostics, especially in third world countries. On the other hand, label-free approaches based on physical characteristics of the cells are often easy to implement, but lack the specificity provided by receptor-ligand interactions4. Control over the transport of cells based on specific receptor-ligand interactions without labeling and label-removal steps would enable cell separation devices for single use or continuous-flow separation, while retaining the specificity of receptor-ligand interactions. In this paper, we wanted to examine whether receptor patterning could be used to achieve nanomechanical control of the transport of cells in two dimensions in a label-free manner through the formation of transient receptor-ligand bonds that result in cell rolling.
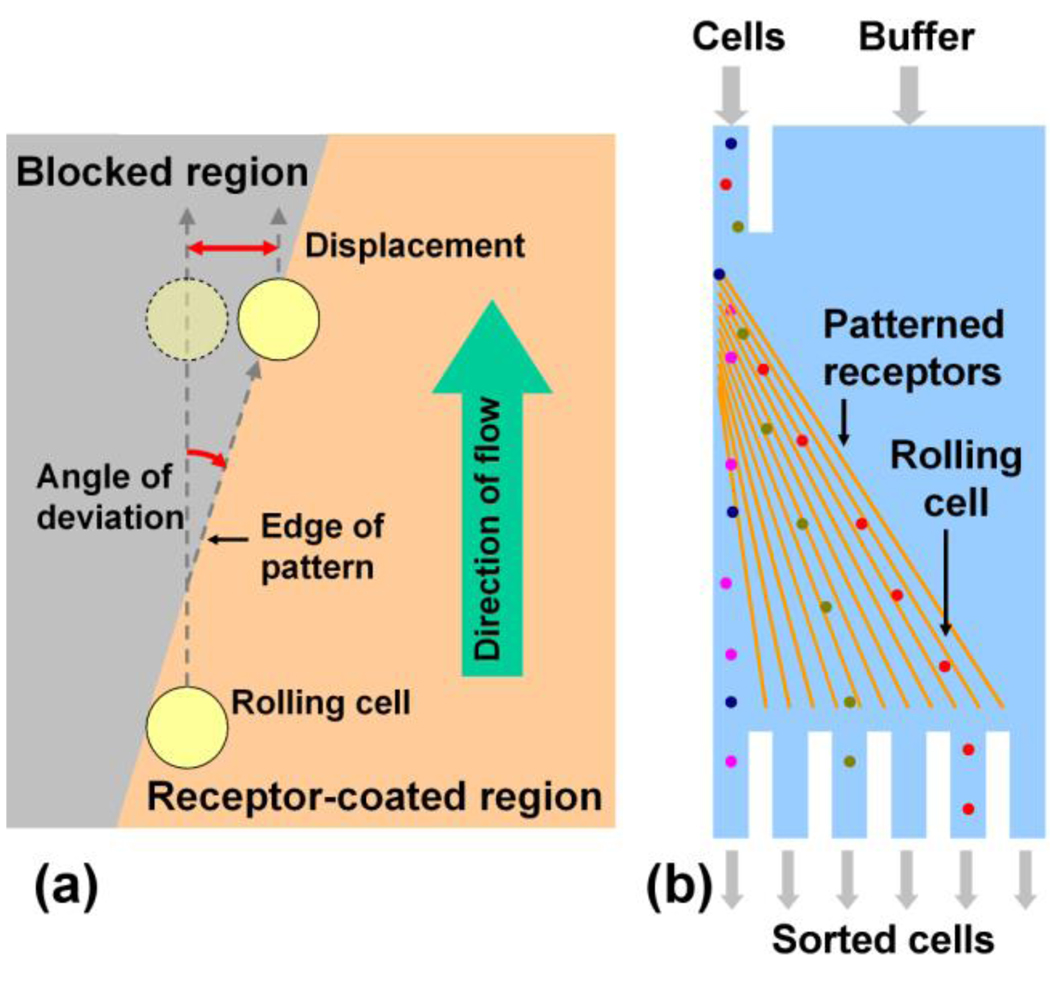
The formation of transient receptor-ligand bonds commonly occurs between cells flowing in the blood stream and the vascular endothelium in a physiological process known as cell rolling6–8. This phenomenon is mediated primarily by glycoprotein receptors known as selectins, among some other receptors can also enable cell rolling8–11. The adhesive bonds formed by selectins have high dissociation rates and are also responsive to shear stress12–14. As the cell rolls under shear force exerted by the flowing blood stream, bonds are formed between the cell and the vascular endothelium on the leading edge of the cell and are broken on the trailing edge of the cell. Leukocytes regulate cell rolling through nanomechanical control via expression of specific adhesion molecules on the surface of specialized 80 nm diameter cell processes known as microvilli that can extend from 350 nm to 1 µm or longer15–18. The number of microvilli tethers during cell rolling has been reported to be quite low, on the order of one to ten15. Cell rolling has been extensively studied due to its important role in physiological processes such as recruitment of leukocytes to sites of inflammation, homing of hematopoietic progenitor cells after intravenous injection, tumor cell metastasis and other inflammatory processes19–21. Cell rolling has been achieved in vitro and mimicked using microsphere models22, 23. Researchers have also investigated the possibility of separating cells based on rolling, including a technique for capturing cells in microfluidic devices24, 25 and separation of cells based on differential rolling velocities in a flow chamber23. Separation based on cell rolling is applicable to a wide range of cell types such as leukocytes, hematopoietic stem/progenitor cells, and metastatic cancer cells19–21, 25. Recent work has also shown that co-immobilization of selectins and antibodies that are specific to molecules expressed on the cell surface can affect the rolling behavior of cells, further enhancing the utility of rolling-based separation26. Current approaches for separation based on cell rolling exploit the rolling behavior to control the transport of cells in one dimension and rely on differential rolling velocities of different cell types: cells that roll faster elute quickly and are thus separated from slowly rolling cells that are either captured or elute slowly. However, this approach requires all cells to start rolling at the same point in time and requires collection of cell fractions at different points in time, necessitating the use of active mechanisms to separate cells into independent compartments. We envisioned that the ability to control cell rolling in two dimensions would enable a simple system that is very easy to implement, and yet retains the specificity due to receptor-ligand interactions (Figure 1b). We therefore explored the possibility of controlling the transport of cells in two dimensions by patterning of selectins on the surface. Herein we demonstrate that edges of P-selectin regions that are at an angle to the direction of fluid flow can be used to direct the motion of rolling cells in two dimensions on the surface (Figure 1a).
Figure 1.
Use of receptor-ligand interactions to direct the motion of cells. (a) Interaction of cells with a receptor-coated substrate may be used to direct the motion of cells along the edge of a receptor pattern. Cells that flow along the direction of fluid flow can be made to follow the receptor edge, and thus be diverted from their direction of flow. (b) This two-dimensional control over the transport of cells may be useful for the design of microfluidic devices for continuous flow separation of cells.
We investigated the effect of a single patterned edge of P-selectin on the motion of rolling HL-60 cells, a human myeloid cell line27 that expresses high levels of P-selectin glycoprotein ligand-1 (PSGL-1) that mediates cell rolling on selectins20. The rolling behavior of HL-60 cells is well-characterized in a number of studies including dependence on shear rate20, 28, cell rigidity and topology29, 30, and capture in a microfluidic device24. HL-60 cells are robust and easy to maintain and also express levels of PSGL-1 that are comparable to leukocytes, making them suitable candidates for our proof-of-concept study.
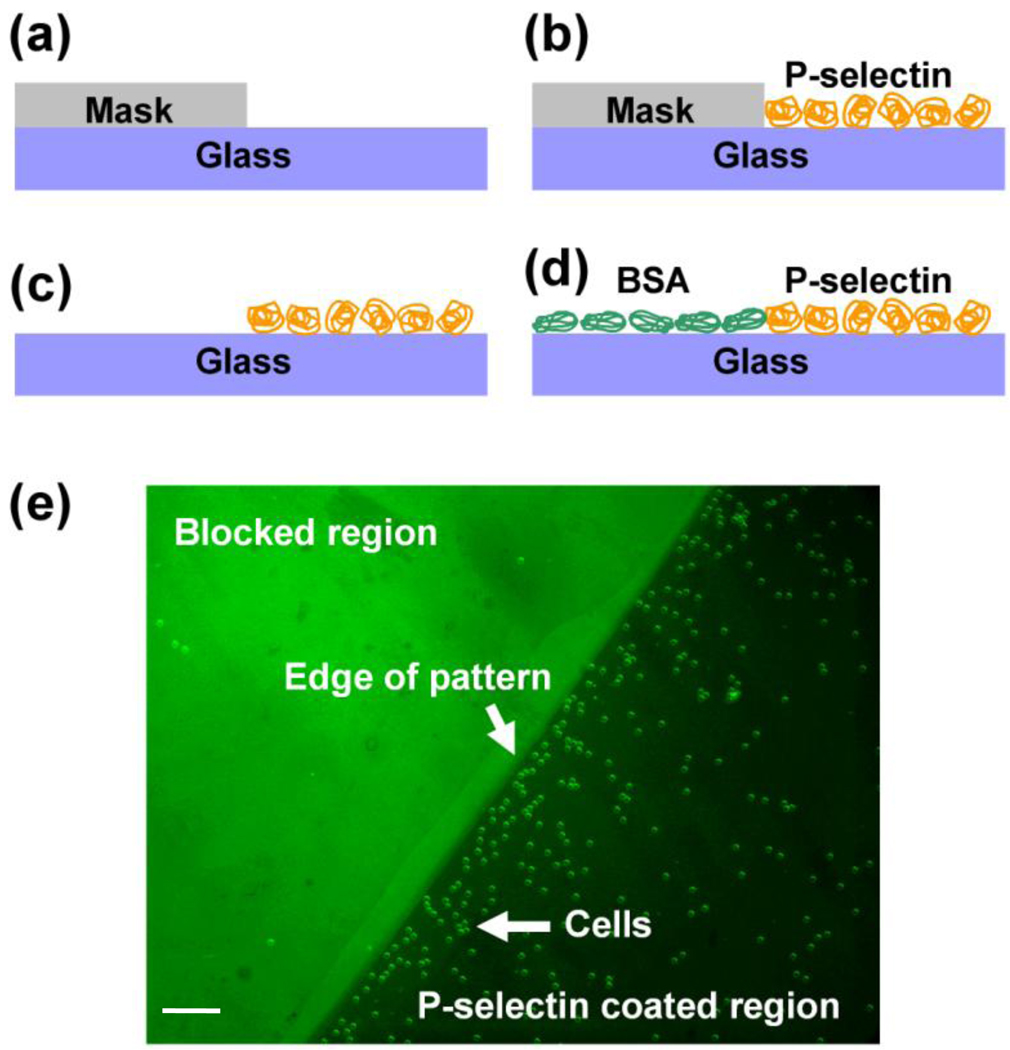
Although covalent immobilization of P-selectin enhances surface properties such as functional stability31, proof of concept studies do not require long term stability and thus physisorption of P-selectin on glass substrates is sufficient. We adopted a simple strategy of selective physisorption of P-selectin by using a silicone rubber mask in order to pattern P-selectin on glass substrates (Figure 2) (see Supporting Information). Use of bovine serum albumin conjugated with fluorescein isothiocyanate (BSA-FITC) during the blocking step revealed selective adsorption of BSA in the region occupied by the silicone mask as compared to the P-selectin coated region. The patterns had well-defined edges, showing that the silicone mask did not leak during the physisorption step. Furthermore, when HL-60 cells were flowed over this patterned substrate the cells selectively interacted with the region coated with P-selectin, confirming the success of the patterning technique (Figure 2e).
Figure 2.
Patterning of P-selectin. A silicone rubber mask was placed on a glass substrate (a) and P-selectin was coated on the exposed area of the substrate by physisorption (b). The silicone mask was then removed from the substrate (c) and BSA was used to block the areas that were not coated with P-selectin (d). Use of fluorescein-labeled BSA enabled visualization of the P-selectin pattern using an epifluorescence microscope (e). HL-60 cells adhered selectively to the P-selectin region, confirming patterning of the substrate with P-selectin. Scale bar: 100 µm.
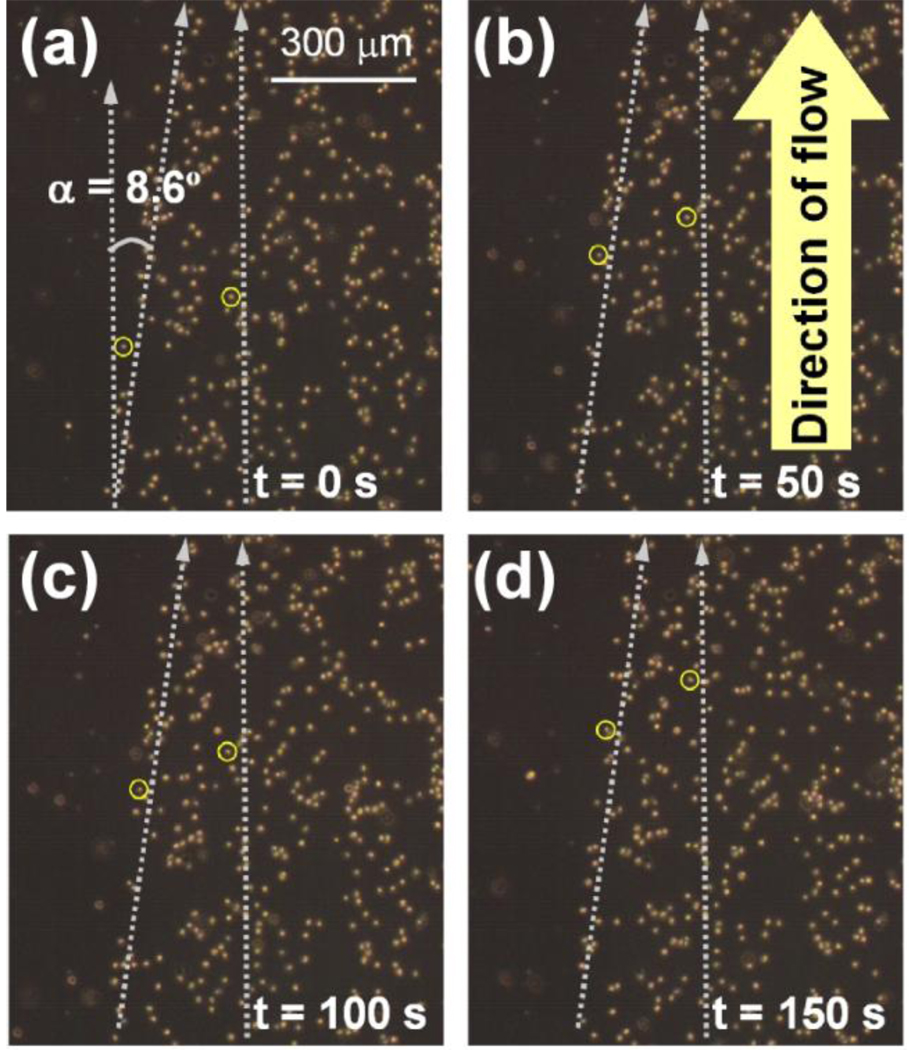
Suspensions of HL-60 cells at densities of 3 – 5 × 105 mL−1 were flowed over the patterned substrates at a shear stress of 0.32 – 12.8 dyn/cm2 (0.03 – 1.28 Pa) using a commercially available flow chamber (see Supporting Information). The flow chamber was rectangular with a width of 1 cm, height of either 125 µm (for cells) or 250 µm (for microspheres), and length of 6 cm, with inlet and outlet at either end. Only some of the cells interacted with the surface, and the remaining cells flowed through the chamber without interacting with the surface; here we are interested in only those cells that interacted with the surface. Selective rolling of HL-60 cells was observed on the P-selectin coated region with slower cell rolling velocities than those on the BSA-coated region where cells were not hindered by the formation of adhesive bonds. Typical velocities of the rolling cells in our experiments ranged from 0.3 to 1.2 µm/s for shear stresses ranging from 0.32 to 12.8 dyn/cm2, which are either comparable to or smaller than cell rolling velocities reported in other studies25, 29, 31, 32. Remarkably, when rolling HL-60 cells encountered the edge of the P-selectin region they were diverted from their original direction of travel along the direction of the edge, demonstrating that receptor patterning could indeed be used to control the transport of cells through transient receptor-ligand adhesive bonds. This effect was observed only for small angles (< c.a. 10 to 15°) between the edge and the direction of flow and nearly all cells that encountered the edge were deflected from their original direction of travel and forced to follow the P-selectin edge. No edge effect was observed at larger edge angles; cells that encountered the edge detached from the substrate and continued to flow in the direction of fluid flow. Thus, the direction of travel of the cells could be changed only at smaller edge angles. Figure 3 shows snapshots of cells rolling under a shear stress of 1.9 dyn/cm2 (300 µL/min), with two cells highlighted- one cell in the P-selectin coated region that did not encounter the edge and another cell that encountered the edge and was forced to travel along the edge. The cell that encountered the edge was deflected from its direction of fluid flow and traveled at an angle of 8.6° with respect to the other cells that did not encounter the edge, demonstrating that a single P-selectin edge could be used to substantially change the direction of cell rolling, and hence control the transport of rolling cells (Videos available in supplementary materials).
Figure 3.
P-selectin edge directs the motion of rolling cells. Rolling HL-60 cells that encountered the edge of a P-selectin pattern making an angle to the fluid flow direction were forced to roll along the edge. The motion of a cell forced to roll along the edge is compared with another cell rolling in the direction of fluid flow, highlighted by circles. The edge succeeded in changing the direction of motion of the rolling cell by 8.6°, resulting in effectively displacing the cell by 0.15 mm from its original position for every 1 mm of length along the direction of flow. Wall shear stress was 1.9 dynes/cm2. Videos are available in Supporting Information.
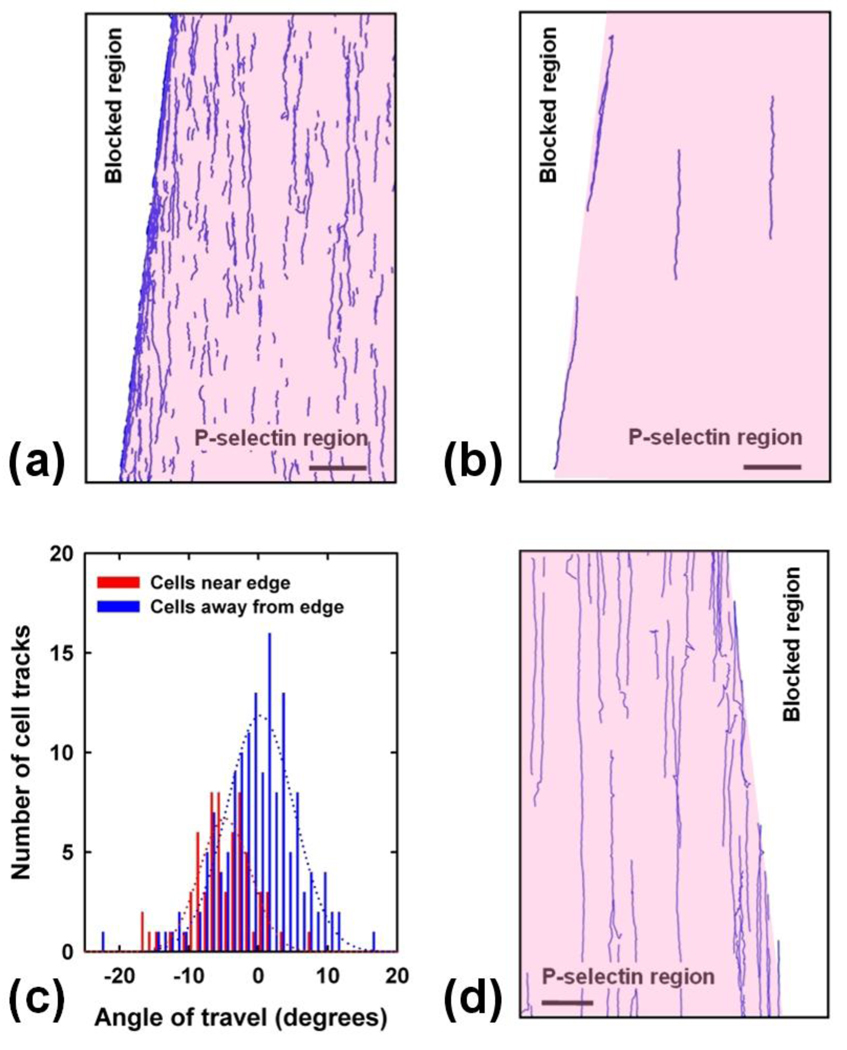
To analyze the rolling behavior of the cells, the sequence of images was processed using Matlab (see Supporting Information). Statically adhered cells were filtered out and tracks of individual cells were plotted, clearly showing the different travel directions of cells rolling on the edge and those rolling inside the P-selectin region (Figure 4a). The image acquisition rate and processing parameters were set so that only those cells that rolled on the surface were tracked. Cells that did not roll moved rapidly as compared to cells that rolled (see supplementary videos), and their large displacements per frame made it impossible to track rolling and free-flowing cells simultaneously. Tracks are not visible in the blocked region as none of the cells rolled in that region. Cells rolling in the P-selectin region that encountered the edge were forced to roll on it instead of crossing over beyond the edge leading to an accumulation of moving cells being transported at an angle to the fluid flow. This effect is evident in the plotted tracks (Figure 4), but not obvious in the images (Figure 3) because of statically adherent cells that accumulated over a period of time. The effect of the P-selectin edge is very clear if only longer cell tracks are plotted (Figure 4b).
Figure 4.
Analysis of cell and microsphere rolling. (a) Matlab tracking of rolling cells generated from a set of 236 images shown in Figure 3 clearly shows the effect of the edge. Inability of cells to cross over the edge resulted in higher density of tracks at the edge. Cell rolling was observed in the P-selectin coated region (pink), but not in the blocked region (white). Scale bar 300 µm. (b) Longer (>300 µm) tracks of cells rolling on the edge and inside the P-selectin region clearly show that the edge affected the rolling direction. Scale bar 300 µm. (c) Angular distribution histogram of the direction of travel of cells rolling near the edge (red) with respect to those away from the edge (blue). Wall shear stress was 1.9 dyn/cm2 (0.19 Pa, 300 µL/min). (d) Similar experiments done with 9.96 µm diameter sLex coated microspheres that roll on P-selectin reveal that the edge did not have a large effect on microspheres as their direction of travel did not change substantially. Wall shear stress was 0.33 dyn/cm2 (0.03 Pa, 200 µL/min). Videos are available in Supporting Information.
To elucidate the effect of the edge on cell rolling, tracks were divided into two sets: (a) Tracks that began within a distance of 30 µm from the edge (cells that encountered the edge), and (b) Tracks that began beyond a distance of 90 µm of the edge (cells that were not influenced by the edge). The direction of travel of each track was identified and plotted as a histogram (Figure 4c), with zero angle corresponding to the mean direction of cells rolling in the P-selectin region. Direction of travel of cells that encountered the edge clearly differed from the mean direction of travel of the other cells by 4–10°. This analysis further confirmed the ability of the edge to control the direction of travel of rolling cells. Furthermore, cells near the edge rolled at an average velocity of 1 µm/s, whereas cells away from the edge rolled at an average velocity of 0.5 µm/s. These results demonstrate that the P-selectin edge enabled control over the transport of rolling cells by (a) changing the direction of rolling, and (b) increasing the rolling speed.
Similar experiments were performed with Sialyl Lewis(x) (sLex) coated microspheres that form transient bonds with P-selectin and are used as models to study cell rolling22, 23, 31. The microspheres rolled selectively on the P-selectin region with average velocity of about 3–4 µm/s at a flow speed of 200 µL/min corresponding to a shear stress of 0.33 dyn/cm2 (0.033 Pa). This velocity is in agreement with our previous work of sLex coated microspheres rolling on P-selectin31. However, the P-selectin edge did not have a significant effect on the direction of rolling of the microspheres: Almost all microspheres that encountered the edge crossed over beyond the edge and their direction of travel remained unchanged (Figure 4d). Tracks corresponding to these microspheres terminated at the edge instead of following it as observed in the case of the rolling cells; once the microspheres detached from the edge, they continued flowing in the direction of fluid flow (see supplementary videos) and did not result in tracks due to their much higher speed.
This observation demonstrates that two types of particles that exhibit similar rolling behavior on P-selectin coated surfaces can exhibit dramatically different rolling behavior on patterned P-selectin. This remarkable difference between the rolling behavior of cells and microspheres at the edge that is not evident in one-dimensional rolling provides insight into rolling at the edge and suggests that the edge effect is capable of differentiating rolling particles based on their nanomechanical properties. We hypothesize that when a cell encounters the edge, an offset between the net force acting on the cell due to fluid flow and forces exerted as the adhesive bonds dissociate causes the cell to undergo asymmetric rolling motion and follow the edge (Figure 5). The moment driving the rolling motion in the direction of fluid flow may be expected to be of the order of Fdrag × a, where a is the radius of the cell or microsphere, and Fdrag is the fluid force acting on the cell. The asymmetric moments that cause the cell to follow the edge may be expected to scale as Fdrag × lcontact, where lcontact is the length scale of the contact area within which the cell or microsphere interact with the substrate. This asymmetric moment may be expected to vanish if the area of contact is very small, since the net force acting on the cell or microsphere would be aligned with the force due to the adhesive bonds, i.e. Fdrag × lcontact would not be large enough to sustain this asymmetric motion, but Fdrag × a would remain relatively unchanged. For the rigid microspheres, the contact length is limited to ~ 0.4 – 0.6 µm assuming that either the bonds or linking molecules can extend by 5 – 10 nm. However, cell rolling depends to a large extent on the mechanical properties of the cell including its deformability and the size and extensibility of microvilli, and the contact length can be several micrometers long for rolling HL-60 cells15, 16, 29. Furthermore, rolling cells can extend long tethers due to extension of microvilli to several micrometers that effectively increases the area of interaction between the rolling cell and the substrate32, 33. Indeed, it is likely to be the reason why sLex coated microspheres selectively rolled on the P-selectin region, but did not follow the edge even when it made a small angle with the direction of fluid flow.
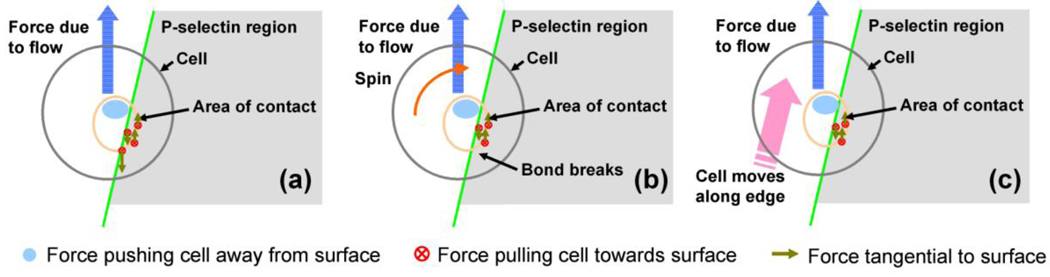
Figure 5.
Potential mechanism of cell rolling along a selectin edge. (a) Bonds on the trailing edge experience maximum strain. When these bonds break, it results in an asymmetric rotation of the cell (b) that causes the cell to move along the edge (c). This mechanism is similar to cell rolling on a surface, but in addition to rotation along an axis parallel to the surface the cell may also spin in plane along an axis perpendicular to the surface as shown in (b). In case of a rigid microsphere, the area of contact is small and the force due to the flow acts through a point vertically above the area of contact, and this asymmetric motion becomes difficult.
Our experiments demonstrate that the transport of cells based on specific receptor-ligand interactions can be controlled in a label-free manner by patterning of receptors that mediate cell rolling. A single edge of P-selectin was capable of substantially changing the trajectory of rolling HL-60 cells with respect to the direction of fluid flow in which the cells would otherwise roll, while affecting rolling microspheres to a much lesser extent. This result suggests the feasibility of developing devices based on nanomechanical control of cell rolling for the creation of simple, cost-effective microfluidic systems for separating a population of cells with slight differences in terms of the rolling behavior. Further characterization of rolling on different surfaces and cell types is essential before cell separation devices based on the edge effect can be developed. Practical cell separation devices will need to ensure that all cells interact with the surface, either by decreasing the height of the flow chamber, by gravity or some other means. Multiple edges will need to be patterned since a single edge may not be sufficient for optimal separation. Since cell rolling is inherently slow, the throughput of such devices will be limited as compared to continuous-flow systems where cell velocities are comparable to the fluid velocity. The operating conditions in these experiments resulted in shear stresses similar to physiological shear stresses experienced by cells in the body. However, higher flow rates may result in shear stresses in large excess of physiological shear stress that may adversely affect the cells. This is a limitation of rolling-based separation, and parallel devices may need to be employed to achieve very high throughput. However, cell separation based on rolling on patterned receptors has the unique advantage in that it can potentially separate cells through specific receptor-ligand interactions in a continuous flow manner without any processing steps. This is a major advantage of this technique as processing steps such as incubation and label removal are expensive and difficult to perform outside the laboratory. For diagnostic purposes, throughput may not be an issue because small volumes of blood may suffice due to the high density of cells. This technique may therefore be suitable in resource-limited settings for analysis of blood where sample processing must be minimized. Rolling on patterned receptors may also find use as a research tool for separation of cells and correlating separated fractions with differences in cell behavior. This unique technique may facilitate the incorporation of factors such as cell deformability, morphology and receptor clustering that influence cell rolling, but are typically inaccessible to conventional techniques of cell separation29, 30, 34, 35, while simultaneously enabling the degree of specificity to be tuned through co-immobilization of antibodies to specific cell markers26. With recent research uncovering the links between nanomechanical characteristics of cells and properties such as states of disease36, 37 and homing ability of cancer cells and stem cells23, the development of separation techniques based on nanomechanical control of cell rolling through patterning of receptors that depends on both the mechanical properties of cells as well as specific receptor-ligand interactions may prove to be a very useful method for separation and analysis of stem cells, cancer cells, and white blood cells.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Minhee Sung for help with the experiments. We also thank the Matlab Particle Tracking Code Repository of Georgetown University and the people who contributed to developing the software. This project was supported by NIH Grant NIH R01 DE016516-01.
REFERENCES
- 1.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304(5673):987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 2.Fu JP, Schoch RB, Stevens AL, Tannenbaum SR, Han JY. Nature Nanotechnology. 2007;2(2):121–128. doi: 10.1038/nnano.2006.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pamme N. Lab on a Chip. 2007;7(12):1644–1659. doi: 10.1039/b712784g. [DOI] [PubMed] [Google Scholar]
- 4.Recktenwald D, Radbruch A, editors. Cell separation methods and applications. New York: M. Dekker; 1998. [Google Scholar]
- 5.Radisic M, Iyer RK, Murthy SK. International Journal of Nanomedicince. 2006;1(1):3–14. doi: 10.2147/nano.2006.1.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granger DN, Kubes P. Journal of Leukocyte Biology. 1994;55(5):662–675. [PubMed] [Google Scholar]
- 7.Lasky LA. Annual Review of Biochemistry. 1995;64:113–139. doi: 10.1146/annurev.bi.64.070195.000553. [DOI] [PubMed] [Google Scholar]
- 8.Tedder TF, Steeber DA, Chen A, Engel P. Faseb Journal. 1995;9(10):866–873. [PubMed] [Google Scholar]
- 9.Mcever RP. Current Opinion in Immunology. 1994;6(1):75–84. doi: 10.1016/0952-7915(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 10.Eniola AO, Krasik EF, Smith LA, Song G, Hammer DA. Biophysical Journal. 2005;89(5):3577–3588. doi: 10.1529/biophysj.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alon R, Kassner PD, Carr MW, Finger EB, Hemler ME, Springer TA. Journal of Cell Biology. 1995;128(6):1243–1253. doi: 10.1083/jcb.128.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell GI. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 13.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Nature. 2003;423(6936):190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 14.Alon R, Hammer DA, Springer TA. Nature. 1995;374(6522):539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 15.Chen SQ, Springer TA. Journal of Cell Biology. 1999;144(1):185–200. doi: 10.1083/jcb.144.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alon R, Chen SQ, Puri KD, Finger EB, Springer TA. Journal of Cell Biology. 1997;138(5):1169–1180. doi: 10.1083/jcb.138.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans E, Heinrich V, Leung A, Kinoshita K. Biophysical Journal. 2005;88(3):2288–2298. doi: 10.1529/biophysj.104.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrich V, Leung A, Evans E. Biophysical Journal. 2005;88(3):2299–2308. doi: 10.1529/biophysj.104.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng JG, Chen M, Chou KC. Current Medicinal Chemistry. 2004;11(16):2153–2160. doi: 10.2174/0929867043364720. [DOI] [PubMed] [Google Scholar]
- 20.Norman KE, Moore KL, Mcever RP, Ley K. Blood. 1995;86(12):4417–4421. [PubMed] [Google Scholar]
- 21.Simon SI, Green CE. Annual Review of Biomedical Engineering. 2005;7:151–185. doi: 10.1146/annurev.bioeng.7.060804.100423. [DOI] [PubMed] [Google Scholar]
- 22.Norman KE, Katopodis AG, Thoma G, Kolbinger F, Hicks AE, Cotter MJ, Pockley AG, Hellewell PG. Blood. 2000;96(10):3585–3591. [PubMed] [Google Scholar]
- 23.Greenberg AW, Hammer DA. Biotechnology and Bioengineering. 2001;73(2):111–124. doi: 10.1002/bit.1043. [DOI] [PubMed] [Google Scholar]
- 24.Chang WC, Lee LP, Liepmann D. Lab on a Chip. 2005;5(1):64–73. doi: 10.1039/b400455h. [DOI] [PubMed] [Google Scholar]
- 25.Narasipura SD, Wojciechowski JC, Charles N, Liesveld JL, King MR. Clinical Chemistry. 2008;54 doi: 10.1373/clinchem.2007.089896. In press. [DOI] [PubMed] [Google Scholar]
- 26.Charles N, Liesveld JL, King MR. Biotechnology Progress. 2007;23(6):1463–1472. doi: 10.1021/bp0702222. [DOI] [PubMed] [Google Scholar]
- 27.Gallagher R, Collins S, Trujillo J, Mccredie K, Ahearn M, Tsai S, Metzgar R, Aulakh G, Ting R, Ruscetti F, Gallo R. Blood. 1979;54(3):713–733. [PubMed] [Google Scholar]
- 28.Lawrence MB, Kansas GS, Kunkel EJ, Ley K. Journal of Cell Biology. 1997;136(3):717–727. doi: 10.1083/jcb.136.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong C, Lei XX. Journal of Biomechanics. 2000;33(1):35–43. doi: 10.1016/s0021-9290(99)00174-8. [DOI] [PubMed] [Google Scholar]
- 30.Wu L, Xiao BT, Jia XL, Zhang Y, Lu SQ, Chen J, Long M. Journal of Biological Chemistry. 2007;282(13):9846–9854. doi: 10.1074/jbc.M609219200. [DOI] [PubMed] [Google Scholar]
- 31.Hong S, Lee D, Zhang H, Zhang JQ, Resvick JN, Khademhosseini A, King MR, Langer R, Karp JM. Langmuir. 2007;23(24):12261–12268. doi: 10.1021/la7014397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran V, Williams M, Yago T, Schmidtke DW, McEver RP. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(37):13519–13524. doi: 10.1073/pnas.0403608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidtke DW, Diamond SL. The Journal of Cell Biology. 2000;149(3):719–729. doi: 10.1083/jcb.149.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus WD, Hochmuth RM. Annals of Biomedical Engineering. 2002;30(10):1273–1280. doi: 10.1114/1.1528614. [DOI] [PubMed] [Google Scholar]
- 35.Caputo KE, Hammer DA. Biophysical Journal. 2005;89(1):187–200. doi: 10.1529/biophysj.104.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nature Nanotechnology. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 37.Suresh S. Acta Materialia. 2007;55(12):3989–4014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.