Figure 3.
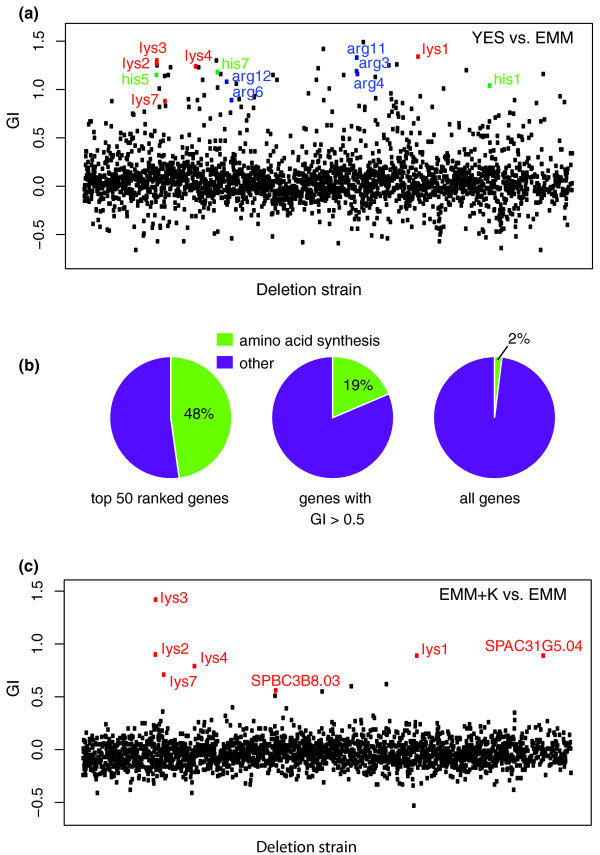
Auxotrophic mutants were revealed by barcode sequencing. (a) The growth inhibition scores (GI) of the deletion mutants grown in rich medium (YES) versus minimal medium (EMM). The strains are ordered on the x-axis according to their positions in the 96-well plates. There are a total of 19 fission yeast genes in the genome database with three-letter names including lys, arg, or his. A calculated GI value is available for 13 of them. These 13 genes whose mutants are known to be auxotrophic for lysine, arginine, or histidine are highlighted in red, blue, and green, respectively. (b) Genes annotated as amino acid biosynthesis genes [GO:0008652] were enriched among the mutants with the highest growth inhibition scores (GI) for YES versus EMM growth conditions. The three pie charts display the percentages of amino acid biosynthesis genes among the genes with the top 50 GI values, among the genes with GI values higher than 0.5, and among all the genes with a GI value. (c) The growth inhibition scores (GI) of the deletion mutants grown in lysine supplemented minimal medium (EMM+K) versus minimal medium (EMM). The seven genes annotated as lysine biosynthesis genes [GO:0009085] are highlighted in red.