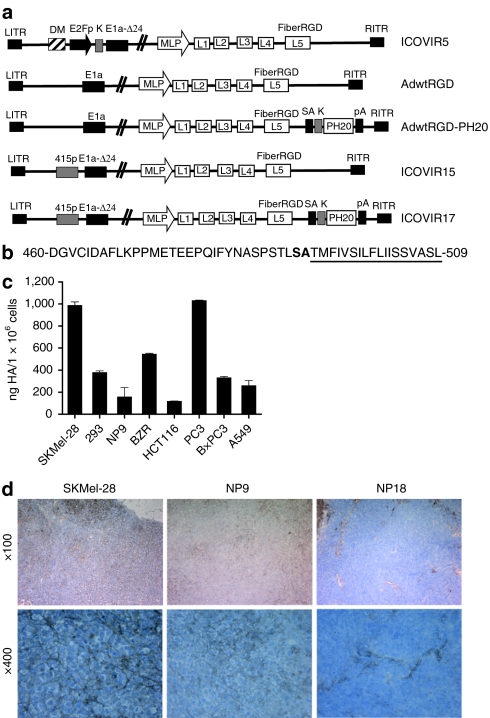
Figure 1.
Adenoviruses and cells used in this study. (a) Schematic representation of viruses used in this study. All viruses contain the RGD peptide in the HI loop of the fiber knob protein, and all oncolytic adenoviruses contain a deletion of 24 base pairs within the E1a region (E1a-Δ24). ICOVIR5 is an oncolytic adenovirus in which the endogenous E1A promoter has been replaced by the human E2F-1 promoter insulated with the DM-1. AdwtRGD and AdwtRGD-PH20 are RGD-modified wild-type adenoviruses. AdwtRGD-PH20 also contains a PH20 cassette inserted downstream of the fiber gene, consisting of a splice acceptor (SA) and a kozak sequence (k) in front of the PH20 cDNA and a polyadenylation sequence (pA). ICOVIR15 and ICOVIR17 are novel oncolytic adenoviruses that contain an E1a promoter modified by the insertion of eight E2F-binding sites and one Sp1-binding site at the nucleotide site 415 of the Ad genome. ICOVIR17 also contains the PH20 cassette after the fiber gene. DM, myotonic dystrophy locus insulator; MLP, major late promoter. (b) Amino acid sequence of the C-terminal human PH20 protein. The hydrophobic region responsible for signal attachment of a GPI anchor is underlined, and the cleavage site is represented in boldface type. The viruses used in this study express a soluble hyaluronidase lacking the cleavage site and the GPI membrane attachment motif. (c) Measurement of HA levels in the supernatants of various tumor cell lines by an ELISA-like assay. Mean ± SD is plotted. (d) Evaluation of HA levels within SKMel-28, NP9 and NP18 tumors by histochemical analysis using the biotinylated hyaluronan-binding protein (HABP-b). HA, hyaluronan.