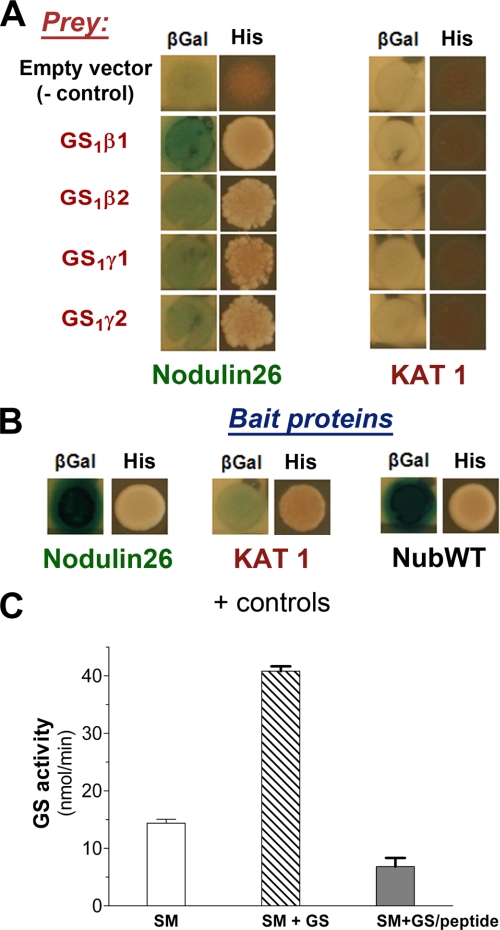
FIGURE 4.
Analysis of interactions between glutamine synthetase and nod26 on SMs and in vivo using the yeast split ubiquitin system. A, S. cerevisiae strains (THY.AP4) containing bait constructs consisting of the nod26 cDNA or the Arabidopsis KAT1 potassium channel cDNA cloned as translational fusions to the C-terminal fragment of ubiquitin (Cub) fused to a synthetic LexA-VP16 transcription factor were mated with THY.AP5 strains containing the GS prey constructs shown. Interaction of bait and prey proteins was tested by activation of two reporter genes; βGal (left panel), shows the results of a β-galactosidase overlay assay, and His (right panel) shows growth on selection medium lacking histidine. The empty vector control shows the results of mating of the indicated bait vectors to the empty pNXgate32 vector. B, the results of positive control matings are shown. In the case of nod26, the mating of homologous bait and prey constructs (resulting in homooligomerization) was performed. A similar mating with homologous KAT1 bait and prey constructs yield positive results because of dimerization (27). NubWT shows the results of using wild-type ubiquitin, which constitutively interacts with Cub fragments and has been used previously as positive control in this system (27). C, isolated purified soybean SMs were incubated with purified native soybean nodule GS (SM + GS) as well as soybean nodule GS preincubated with 10 μm nod26 peptide (SM+GS/peptide). As a control, SMs were incubated with an equivalent volume of binding buffer without added GS (SM). Membranes were washed, and the GS activity bound was assayed with the error bars showing S.E. (n = 6).