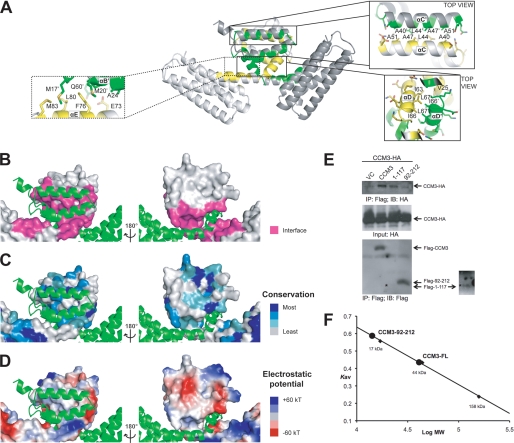
FIGURE 2.
CCM3 dimerization. A, scheme representation of the CCM3 dimer. Exploded views show the anti-parallel hydrophobic stripe for helix αC and the symmetric hydrophobic focal point formed by Ile-66 and Leu-67. B, interacting residues. Surfaces are colored pink for residues that mediate the N-terminal dimerization interface. C, sequence conservation. The surface colored by sequence conservation is based on 34 CCM3 sequences. Darker blue indicates higher conservation. Generated using Consurf (36). D, electrostatic potential representation (+60 kT, blue; −60 kT, red). E, immunoprecipitation. CCM3-HA was co-transfected with Flag-CCM3 variants as indicated into 293T cells. Association of CCM3-HA with full-length CCM3, CCM3 N-terminal region (1–117), and CCM3 FAT domain (92–212) was determined by IP with anti-Flag (M2) followed by Western blot with anti-HA. Flag-CCM3 in the immunoprecipitates was determined by Western blot with anti-Flag. CCM3-HA in the input was also determined. Arrows indicate CCM3. Flag-1–117 is indicated with an (*) and shown in the overexposed panel. VC, vector control. F, size exclusion chromatography. A Superdex 200 analytical grade SEC column was used to analyze CCM3 constructs 1–212 and 92–212. Full-length (1–212) CCM3 elutes as a dimer (predicted mass: ∼25 kDa) and construct 92–212 elutes as a monomer (predicted mass: ∼14 kDa). Standards used to calibrate the column were bovine γ-globulin 158 kDa, chicken ovalbumin 44 kDa, and equine myoglobin 17 kDa. Kav = (Ve−Vo)/(Vt−Vo) where Ve is the elution volume of the protein, Vo is the void volume, and Vt is the total bead volume.