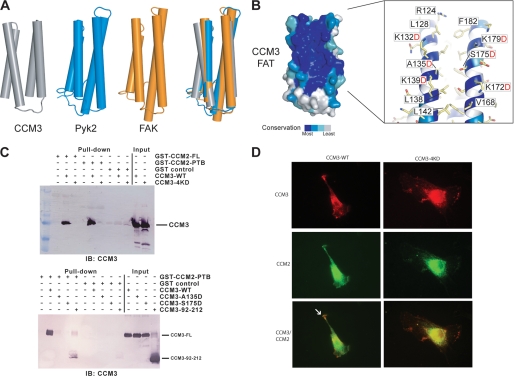
FIGURE 3.
CCM3 FAT-homology domain binds CCM2. A, superposition of CCM3, Pyk2, and FAK. The four helices of Pyk2 FAT domain in blue (PDB ID: 3GM1) (22) and FAK FAT domain in orange (PDB ID: 1OW7) (23) are superposed with the FAT-homology domain of CCM3 (gray). B, surface conservation. The CCM3 FAT-homology domain shows very high conservation on the surfaces of helices αG and αH (see alignment in supplemental Fig. S3). The exploded view labels some of the highly conserved residues and those mutated in this study. C, pull-down. Top: GST fusions of both full-length CCM2 and the predicted CCM2-PTB domain pull-down full-length CCM3. Quadruple mutation of conserved lysines in CCM3, K132D, K139D, K172D, and K179D (CCM3–4KD) renders CCM3 unable to pull-down with CCM2. Bottom: CCM3 HP1 mutations A135D and S175D inhibit pull-down of CCM3 with CCM2-PTB domain. Both full-length CCM3 and CCM3–92-212 pull-down with CCM2-PTB. CCM3 constructs were purified by size exclusion chromatography and eluted as monodisperse peaks. D, immunofluorescence. Either CCM3 FAT-homology domain (lhs) or CCM3 FAT-homology-4KD (rhs) were co-transfected into BAECs with CCM2 and detected by indirect immunofluorescence. CCM2 and CCM3 FAT-homology partially co-localize (white arrow), but introduction of HP1 mutations reduce this co-localization. CCM3 constructs included a C-terminal Flag tag and CCM2, an N-terminal GFP tag.