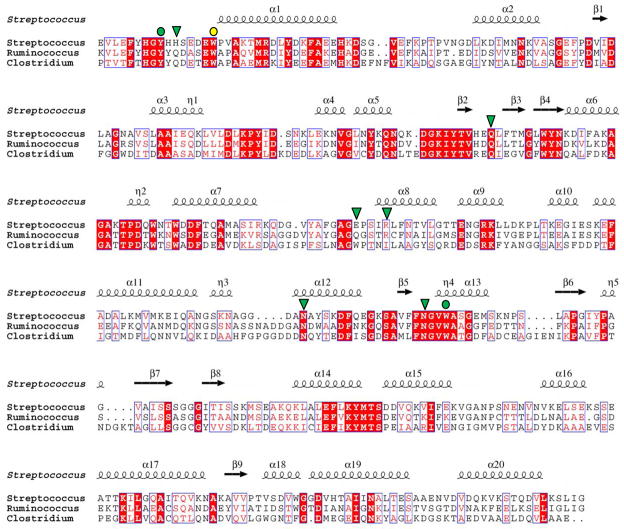
Figure 4.
Alignment of FcsSBP with its most closely related proteins in the database. Streptococcus denotes FcsSBP from Streptococcus pneumoniae SP3-BS71, Ruminococcus denotes RUMGNA_03829 from Ruminococcus gnavus (ATCC 29149), Clostridium denotes CLOSCI_02090 from Clostridium scindens (ATCC 35704). The secondary structure of FcsSBP is indicated above the sequence. Residues in FcsSBP that are involved in hydrogen bonding with substrate are indicated above the alignment by green shapes (green triangle, hydrogen bonding only; green circle, apolar and hydrogen bonding interaction). Aromatic residues involved in apolar interactions are indicated by circles (yellow circle, apolar interaction only; green circle, apolar and hydrogen bonding interaction).